+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human liver mitochondrial Glutamate dehydrogenase 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | human / liver / mitochondrial / Glutamate dehydrogenase 1 / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationL-glutamate dehydrogenase [NAD(P)+] activity / L-leucine binding / tricarboxylic acid metabolic process / glutamate dehydrogenase [NAD(P)+] / : / L-glutamate dehydrogenase (NADP+) activity / L-glutamate dehydrogenase (NAD+) activity / Glutamate and glutamine metabolism / L-glutamate catabolic process / glutamine metabolic process ...L-glutamate dehydrogenase [NAD(P)+] activity / L-leucine binding / tricarboxylic acid metabolic process / glutamate dehydrogenase [NAD(P)+] / : / L-glutamate dehydrogenase (NADP+) activity / L-glutamate dehydrogenase (NAD+) activity / Glutamate and glutamine metabolism / L-glutamate catabolic process / glutamine metabolic process / NAD+ binding / substantia nigra development / Mitochondrial protein degradation / Transcriptional activation of mitochondrial biogenesis / positive regulation of insulin secretion / ADP binding / mitochondrial matrix / GTP binding / endoplasmic reticulum / protein homodimerization activity / mitochondrion / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.31 Å | |||||||||
 Authors Authors | Zhang Z / Tringides M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell Proteomics / Year: 2023 Journal: Mol Cell Proteomics / Year: 2023Title: High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology. Authors: Zhemin Zhang / Marios L Tringides / Christopher E Morgan / Masaru Miyagi / Jason A Mears / Charles L Hoppel / Edward W Yu /  Abstract: The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of ...The application of integrated systems biology to the field of structural biology is a promising new direction, although it is still in the infant stages of development. Here we report the use of single particle cryo-EM to identify multiple proteins from three enriched heterogeneous fractions prepared from human liver mitochondrial lysate. We simultaneously identify and solve high-resolution structures of nine essential mitochondrial enzymes with key metabolic functions, including fatty acid catabolism, reactive oxidative species clearance, and amino acid metabolism. Our methodology also identified multiple distinct members of the acyl-CoA dehydrogenase family. This work highlights the potential of cryo-EM to explore tissue proteomics at the atomic level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40558.map.gz emd_40558.map.gz | 51.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40558-v30.xml emd-40558-v30.xml emd-40558.xml emd-40558.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40558_fsc.xml emd_40558_fsc.xml | 11 KB | Display |  FSC data file FSC data file |
| Images |  emd_40558.png emd_40558.png | 143 KB | ||
| Filedesc metadata |  emd-40558.cif.gz emd-40558.cif.gz | 5.5 KB | ||
| Others |  emd_40558_half_map_1.map.gz emd_40558_half_map_1.map.gz emd_40558_half_map_2.map.gz emd_40558_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40558 http://ftp.pdbj.org/pub/emdb/structures/EMD-40558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40558 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40558 | HTTPS FTP |
-Related structure data
| Related structure data |  8sk8MC  8sgpC  8sgrC  8sgsC  8sgvC  8shsC  8sk6C  8skrC  8sksC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40558.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40558.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
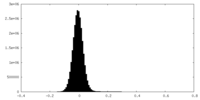
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_40558_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
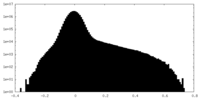
| Density Histograms |
-Half map: #2
| File | emd_40558_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
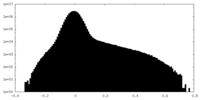
| Density Histograms |
- Sample components
Sample components
-Entire : Glutamate dehydrogenase 1
| Entire | Name: Glutamate dehydrogenase 1 |
|---|---|
| Components |
|
-Supramolecule #1: Glutamate dehydrogenase 1
| Supramolecule | Name: Glutamate dehydrogenase 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glutamate dehydrogenase 1, mitochondrial
| Macromolecule | Name: Glutamate dehydrogenase 1, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: glutamate dehydrogenase [NAD(P)+] |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.480746 KDa |
| Sequence | String: MYRYLGEALL LSRAGPAALG SASADSAALL GWARGQPAAA PQPGLALAAR RHYSEAVADR EDDPNFFKMV EGFFDRGASI VEDKLVEDL RTRESEEQKR NRVRGILRII KPCNHVLSLS FPIRRDDGSW EVIEGYRAQH SQHRTPCKGG IRYSTDVSVD E VKALASLM ...String: MYRYLGEALL LSRAGPAALG SASADSAALL GWARGQPAAA PQPGLALAAR RHYSEAVADR EDDPNFFKMV EGFFDRGASI VEDKLVEDL RTRESEEQKR NRVRGILRII KPCNHVLSLS FPIRRDDGSW EVIEGYRAQH SQHRTPCKGG IRYSTDVSVD E VKALASLM TYKCAVVDVP FGGAKAGVKI NPKNYTDNEL EKITRRFTME LAKKGFIGPG IDVPAPDMST GEREMSWIAD TY ASTIGHY DINAHACVTG KPISQGGIHG RISATGRGVF HGIENFINEA SYMSILGMTP GFGDKTFVVQ GFGNVGLHSM RYL HRFGAK CIAVGESDGS IWNPDGIDPK ELEDFKLQHG SILGFPKAKP YEGSILEADC DILIPAASEK QLTKSNAPRV KAKI IAEGA NGPTTPEADK IFLERNIMVI PDLYLNAGGV TVSYFEWLKN LNHVSYGRLT FKYERDSNYH LLMSVQESLE RKFGK HGGT IPIVPTAEFQ DRISGASEKD IVHSGLAYTM ERSARQIMRT AMKYNLGLDL RTAAYVNAIE KVFKVYNEAG VTFT UniProtKB: Glutamate dehydrogenase 1, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
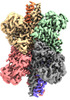
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)