[English] 日本語
 Yorodumi
Yorodumi- EMDB-4045: 13-protofilament microtubule structure determined in situ from U2... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4045 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 13-protofilament microtubule structure determined in situ from U2OS cell | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
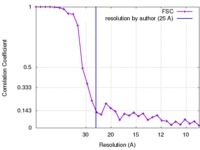
| Method | subtomogram averaging / cryo EM / Resolution: 25.0 Å | |||||||||
 Authors Authors | Grange M / Vasishtan D / Gruenewald K | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2017 Journal: J Struct Biol / Year: 2017Title: Cellular electron cryo tomography and in situ sub-volume averaging reveal the context of microtubule-based processes. Authors: Michael Grange / Daven Vasishtan / Kay Grünewald /  Abstract: Electron cryo-tomography (cryoET) is currently the only technique that allows the direct observation of proteins in their native cellular environment. Sub-volume averaging of electron tomograms ...Electron cryo-tomography (cryoET) is currently the only technique that allows the direct observation of proteins in their native cellular environment. Sub-volume averaging of electron tomograms offers a route to increase the signal-to-noise of repetitive biological structures, such improving the information content and interpretability of tomograms. We discuss the potential for sub-volume averaging in highlighting and investigating specific processes in situ, focusing on microtubule structure and viral infection. We show that (i) in situ sub-volume averaging from single tomograms can guide and complement segmentation of biological features, (ii) the in situ determination of the structure of individual viruses is possible as they infect a cell, and (iii) novel, transient processes can be imaged with high levels of detail. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4045.map.gz emd_4045.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4045-v30.xml emd-4045-v30.xml emd-4045.xml emd-4045.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4045_fsc.xml emd_4045_fsc.xml | 3 KB | Display |  FSC data file FSC data file |
| Images |  emd_4045.png emd_4045.png | 106.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4045 http://ftp.pdbj.org/pub/emdb/structures/EMD-4045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4045 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4045.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4045.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
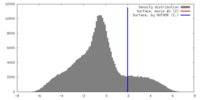
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Microtubule
| Entire | Name: Microtubule |
|---|---|
| Components |
|
-Supramolecule #1: Microtubule
| Supramolecule | Name: Microtubule / type: cell / ID: 1 / Parent: 0 / Details: microtubule from U2OS |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: U2OS / Tissue: Cell Homo sapiens (human) / Strain: U2OS / Tissue: Cell |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: Cell |
|---|---|
| Grid | Model: C-flat 2/2 holey carbon gold supported grids (EMS) / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 60 % / Chamber temperature: 298 K / Instrument: HOMEMADE PLUNGER Details: Adherent cells grown on electron microscopy grid, manually blotted for 3s and plunge frozen. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 77.0 K / Max: 110.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3836 pixel / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 5.0 µm / Calibrated defocus min: 5.0 µm / Calibrated magnification: 21739 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 95000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)