+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Adenosylcobalamin-bound riboswitch dimer, form 3 | |||||||||
 Map data Map data | unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA cobalamin riboswitch / RNA | |||||||||
| Biological species |  Caldanaerobacter subterraneus subsp. tengcongensis (bacteria) Caldanaerobacter subterraneus subsp. tengcongensis (bacteria) | |||||||||
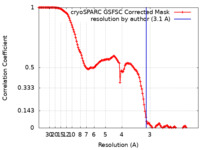
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Ding J / Deme JC / Stagno JR / Yu P / Lea SM / Wang YX | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM. Authors: Jienyu Ding / Justin C Deme / Jason R Stagno / Ping Yu / Susan M Lea / Yun-Xing Wang /  Abstract: RNA conformational heterogeneity often hampers its high-resolution structure determination, especially for large and flexible RNAs devoid of stabilizing proteins or ligands. The adenosylcobalamin ...RNA conformational heterogeneity often hampers its high-resolution structure determination, especially for large and flexible RNAs devoid of stabilizing proteins or ligands. The adenosylcobalamin riboswitch exhibits heterogeneous conformations under 1 mM Mg2+ concentration and ligand binding reduces conformational flexibility. Among all conformers, we determined one apo (5.3 Å) and four holo cryo-electron microscopy structures (overall 3.0-3.5 Å, binding pocket 2.9-3.2 Å). The holo dimers exhibit global motions of helical twisting and bending around the dimer interface. A backbone comparison of the apo and holo states reveals a large structural difference in the P6 extension position. The central strand of the binding pocket, junction 6/3, changes from an 'S'- to a 'U'-shaped conformation to accommodate ligand. Furthermore, the binding pocket can partially form under 1 mM Mg2+ and fully form under 10 mM Mg2+ within the bound-like structure in the absence of ligand. Our results not only demonstrate the stabilizing ligand-induced conformational changes in and around the binding pocket but may also provide further insight into the role of the P6 extension in ligand binding and selectivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40264.map.gz emd_40264.map.gz | 217.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40264-v30.xml emd-40264-v30.xml emd-40264.xml emd-40264.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40264_fsc.xml emd_40264_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40264.png emd_40264.png | 26.4 KB | ||
| Masks |  emd_40264_msk_1.map emd_40264_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40264.cif.gz emd-40264.cif.gz | 5.5 KB | ||
| Others |  emd_40264_additional_1.map.gz emd_40264_additional_1.map.gz emd_40264_half_map_1.map.gz emd_40264_half_map_1.map.gz emd_40264_half_map_2.map.gz emd_40264_half_map_2.map.gz | 230.3 MB 226.2 MB 226.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40264 http://ftp.pdbj.org/pub/emdb/structures/EMD-40264 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40264 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40264 | HTTPS FTP |
-Related structure data
| Related structure data |  8sa4MC  8sa2C  8sa3C  8sa5C  8sa6C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40264.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40264.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0395 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40264_msk_1.map emd_40264_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: B-factor sharpened map
| File | emd_40264_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_40264_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_40264_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : holodimer 3
| Entire | Name: holodimer 3 |
|---|---|
| Components |
|
-Supramolecule #1: holodimer 3
| Supramolecule | Name: holodimer 3 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Caldanaerobacter subterraneus subsp. tengcongensis (bacteria) Caldanaerobacter subterraneus subsp. tengcongensis (bacteria) |
-Macromolecule #1: adenosylcobalamin riboswitch form 3
| Macromolecule | Name: adenosylcobalamin riboswitch form 3 / type: rna / ID: 1 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  Caldanaerobacter subterraneus subsp. tengcongensis (bacteria) Caldanaerobacter subterraneus subsp. tengcongensis (bacteria) |
| Molecular weight | Theoretical: 68.273664 KDa |
| Sequence | String: GGUUAAAGCC UUAUGGUCGC UACCAUUGCA CUCCGGUAGC GUUAAAAGGG AAGACGGGUG AGAAUCCCGC GCAGCCCCCG CUACUGUGA GGGAGGACGA AGCCCUAGUA AGCCACUGCC GAAAGGUGGG AAGGCAGGGU GGAGGAUGAG UCCCGAGCCA G GAGACCUG ...String: GGUUAAAGCC UUAUGGUCGC UACCAUUGCA CUCCGGUAGC GUUAAAAGGG AAGACGGGUG AGAAUCCCGC GCAGCCCCCG CUACUGUGA GGGAGGACGA AGCCCUAGUA AGCCACUGCC GAAAGGUGGG AAGGCAGGGU GGAGGAUGAG UCCCGAGCCA G GAGACCUG CCAUAAGGUU UUAGAAGUUC GCCUUCGGGG GGAAGGUGAA CA |
-Macromolecule #2: Adenosylcobalamin
| Macromolecule | Name: Adenosylcobalamin / type: ligand / ID: 2 / Number of copies: 1 / Formula: B1Z |
|---|---|
| Molecular weight | Theoretical: 1.58059 KDa |
| Chemical component information | 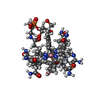 ChemComp-B1Z: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 54.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)