+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human ER membrane protein complex (EMC) in GDN, 8-subunit map | |||||||||
 Map data Map data | Full map file | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Insertase / endoplasmic reticulum / transmembrane chaperone / MEMBRANE PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Tomaleri GP / Nguyen V / Januszyk K / Voorhees RM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Cell Biol / Year: 2023 Journal: J Cell Biol / Year: 2023Title: A selectivity filter in the ER membrane protein complex limits protein misinsertion at the ER. Authors: Tino Pleiner / Masami Hazu / Giovani Pinton Tomaleri / Vy N Nguyen / Kurt Januszyk / Rebecca M Voorhees /  Abstract: Tail-anchored (TA) proteins play essential roles in mammalian cells, and their accurate localization is critical for proteostasis. Biophysical similarities lead to mistargeting of mitochondrial TA ...Tail-anchored (TA) proteins play essential roles in mammalian cells, and their accurate localization is critical for proteostasis. Biophysical similarities lead to mistargeting of mitochondrial TA proteins to the ER, where they are delivered to the insertase, the ER membrane protein complex (EMC). Leveraging an improved structural model of the human EMC, we used mutagenesis and site-specific crosslinking to map the path of a TA protein from its cytosolic capture by methionine-rich loops to its membrane insertion through a hydrophilic vestibule. Positively charged residues at the entrance to the vestibule function as a selectivity filter that uses charge-repulsion to reject mitochondrial TA proteins. Similarly, this selectivity filter retains the positively charged soluble domains of multipass substrates in the cytosol, thereby ensuring they adopt the correct topology and enforcing the "positive-inside" rule. Substrate discrimination by the EMC provides a biochemical explanation for one role of charge in TA protein sorting and protects compartment integrity by limiting protein misinsertion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40247.map.gz emd_40247.map.gz | 121.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40247-v30.xml emd-40247-v30.xml emd-40247.xml emd-40247.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40247.png emd_40247.png | 51.1 KB | ||
| Others |  emd_40247_additional_1.map.gz emd_40247_additional_1.map.gz emd_40247_half_map_1.map.gz emd_40247_half_map_1.map.gz emd_40247_half_map_2.map.gz emd_40247_half_map_2.map.gz | 124.5 MB 226.4 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40247 http://ftp.pdbj.org/pub/emdb/structures/EMD-40247 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40247 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40247 | HTTPS FTP |
-Validation report
| Summary document |  emd_40247_validation.pdf.gz emd_40247_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40247_full_validation.pdf.gz emd_40247_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_40247_validation.xml.gz emd_40247_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_40247_validation.cif.gz emd_40247_validation.cif.gz | 18.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40247 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40247 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40247 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40247 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40247.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40247.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map file | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Sharped Full map file
| File | emd_40247_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharped Full map file | ||||||||||||
| Projections & Slices |
| ||||||||||||
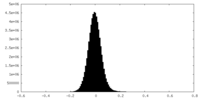
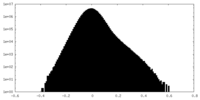
| Density Histograms |
-Half map: Half map (A)
| File | emd_40247_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (A) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map (B)
| File | emd_40247_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (B) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human ER Membrane Protein Complex
| Entire | Name: Human ER Membrane Protein Complex |
|---|---|
| Components |
|
-Supramolecule #1: Human ER Membrane Protein Complex
| Supramolecule | Name: Human ER Membrane Protein Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Cell: HEK293 Homo sapiens (human) / Cell: HEK293 |
-Macromolecule #1: ER membrane protein complex subunit 1
| Macromolecule | Name: ER membrane protein complex subunit 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAAEWASRFW LWATLLIPAA AVYEDQVGKF DWRQQYVGKV KFASLEFSPG SKKLVVATEK NVIAALNSRT GEILWRHVDK GTAEGAVDA MLLHGQDVIT VSNGGRIMRS WETNIGGLNW EITLDSGSFQ ALGLVGLQES VRYIAVLKKT TLALHHLSSG H LKWVEHLP ...String: MAAEWASRFW LWATLLIPAA AVYEDQVGKF DWRQQYVGKV KFASLEFSPG SKKLVVATEK NVIAALNSRT GEILWRHVDK GTAEGAVDA MLLHGQDVIT VSNGGRIMRS WETNIGGLNW EITLDSGSFQ ALGLVGLQES VRYIAVLKKT TLALHHLSSG H LKWVEHLP ESDSIHYQMV YSYGSGVVWA LGVVPFSHVN IVKFNVEDGE IVQQVRVSTP WLQHLSGACG VVDEAVLVCP DP SSRSLQT LALETEWELR QIPLQSLDLE FGSGFQPRVL PTQPNPVDAS RAQFFLHLSP SHYALLQYHY GTLSLLKNFP QTA LVSFAT TGEKTVAAVM ACRNEVQKSS SSEDGSMGSF SEKSSSKDSL ACFNQTYTIN LYLVETGRRL LDTTITFSLE QSGT RPERL YIQVFLKKDD SVGYRALVQT EDHLLLFLQQ LAGKVVLWSR EESLAEVVCL EMVDLPLTGA QAELEGEFGK KADGL LGMF LKRLSSQLIL LQAWTSHLWK MFYDARKPRS QIKNEINIDT LARDEFNLQK MMVMVTASGK LFGIESSSGT ILWKQY LPN VKPDSSFKLM VQRTTAHFPH PPQCTLLVKD KESGMSSLYV FNPIFGKWSQ VAPPVLKRPI LQSLLLPVMD QDYAKVL LL IDDEYKVTAF PATRNVLRQL HELAPSIFFY LVDAEQGRLC GYRLRKDLTT ELSWELTIPP EVQRIVKVKG KRSSEHVH S QGRVMGDRSV LYKSLNPNLL AVVTESTDAH HERTFIGIFL IDGVTGRIIH SSVQKKAKGP VHIVHSENWV VYQYWNTKA RRNEFTVLEL YEGTEQYNAT AFSSLDRPQL PQVLQQSYIF PSSISAMEAT ITERGITSRH LLIGLPSGAI LSLPKALLDP RRPEIPTEQ SREENLIPYS PDVQIHAERF INYNQTVSRM RGIYTAPSGL ESTCLVVAYG LDIYQTRVYP SKQFDVLKDD Y DYVLISSV LFGLVFATMI TKRLAQVKLL NRAWR |
-Macromolecule #2: ER membrane protein complex subunit 2
| Macromolecule | Name: ER membrane protein complex subunit 2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAKVSELYDV TWEEMRDKMR KWREENSRNS EQIVEVGEEL INEYASKLGD DIWIIYEQVM IAALDYGRDD LALFCLQELR RQFPGSHRV KRLTGMRFEA MERYDDAIQL YDRILQEDPT NTAARKRKIA IRKAQGKNVE AIRELNEYLE QFVGDQEAWH E LAELYINE ...String: MAKVSELYDV TWEEMRDKMR KWREENSRNS EQIVEVGEEL INEYASKLGD DIWIIYEQVM IAALDYGRDD LALFCLQELR RQFPGSHRV KRLTGMRFEA MERYDDAIQL YDRILQEDPT NTAARKRKIA IRKAQGKNVE AIRELNEYLE QFVGDQEAWH E LAELYINE HDYAKAAFCL EELMMTNPHN HLYCQQYAEV KYTQGGLENL ELSRKYFAQA LKLNNRNMRA LFGLYMSASH IA SNPKASA KTKKDNMKYA SWAASQINRA YQFAGRSKKE TKYSLKAVED MLETLQITQS |
-Macromolecule #3: ER membrane protein complex subunit 3
| Macromolecule | Name: ER membrane protein complex subunit 3 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAGPELLLDS NIRLWVVLPI VIITFFVGMI RHYVSILLQS DKKLTQEQVS DSQVLIRSRV LRENGKYIPK QSFLTRKYYF NNPEDGFFK KTKRKVVPPS PMTDPTMLTD MMKGNVTNVL PMILIGGWIN MTFSGFVTTK VPFPLTLRFK PMLQQGIELL T LDASWVSS ...String: MAGPELLLDS NIRLWVVLPI VIITFFVGMI RHYVSILLQS DKKLTQEQVS DSQVLIRSRV LRENGKYIPK QSFLTRKYYF NNPEDGFFK KTKRKVVPPS PMTDPTMLTD MMKGNVTNVL PMILIGGWIN MTFSGFVTTK VPFPLTLRFK PMLQQGIELL T LDASWVSS ASWYFLNVFG LRSIYSLILG QDNAADQSRM MQEQMTGAAM AMPADTNKAF KTEWEALELT DHQWALDDVE EE LMAKDLH FEGMFKKELQ TSIF |
-Macromolecule #4: ER membrane protein complex subunit 4
| Macromolecule | Name: ER membrane protein complex subunit 4 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MTAQGGLVAN RGRRFKWAIE LSGPGGGSRG RSDRGSGQGD SLYPVGYLDK QVPDTSVQET DRILVEKRCW DIALGPLKQI PMNLFIMYM AGNTISIFPT MMVCMMAWRP IQALMAISAT FKMLESSSQK FLQGLVYLIG NLMGLALAVY KCQSMGLLPT H ASDWLAFI EPPERMEFSG GGLLL |
-Macromolecule #5: ER membrane protein complex subunit 5
| Macromolecule | Name: ER membrane protein complex subunit 5 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAPSLWKGLV GIGLFALAHA AFSAAQHRSY MRLTEKEDES LPIDIVLQTL LAFAVTCYGI VHIAGEFKDM DATSELKNKT FDTLRNHPS FYVFNHRGRV LFRPSDTANS SNQDALSSNT SLKLRKLESL RR |
-Macromolecule #6: ER membrane protein complex subunit 6
| Macromolecule | Name: ER membrane protein complex subunit 6 / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAAVVAKREG PPFISEAAVR GNAAVLDYCR TSVSALSGAT AGILGLTGLY GFIFYLLASV LLSLLLILKA GRRWNKYFKS RRPLFTGGL IGGLFTYVLF WTFLYGMVHV Y |
-Macromolecule #7: ER membrane protein complex subunit 7
| Macromolecule | Name: ER membrane protein complex subunit 7 / type: protein_or_peptide / ID: 7 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAAALWGFFP VLLLLLLSGD VQSSEVPGAA AEGSGGSGVG IGDRFKIEGR AVVPGVKPQD WISAARVLVD GEEHVGFLKT DGSFVVHDI PSGSYVVEVV SPAYRFDPVR VDITSKGKMR ARYVNYIKTS EVVRLPYPLQ MKSSGPPSYF IKRESWGWTD F LMNPMVMM ...String: MAAALWGFFP VLLLLLLSGD VQSSEVPGAA AEGSGGSGVG IGDRFKIEGR AVVPGVKPQD WISAARVLVD GEEHVGFLKT DGSFVVHDI PSGSYVVEVV SPAYRFDPVR VDITSKGKMR ARYVNYIKTS EVVRLPYPLQ MKSSGPPSYF IKRESWGWTD F LMNPMVMM MVLPLLIFVL LPKVVNTSDP DMRREMEQSM NMLNSNHELP DVSEFMTRLF SSKSSGKSSS GSSKTGKSGA GK RR |
-Macromolecule #8: ER membrane protein complex subunit 8
| Macromolecule | Name: ER membrane protein complex subunit 8 / type: protein_or_peptide / ID: 8 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MPGVKLTTQA YCKMVLHGAK YPHCAVNGLL VAEKQKPRKE HLPLGGPGAH HTLFVDCIPL FHGTLALAPM LEVALTLIDS WCKDHSYVI AGYYQANERV KDASPNQVAE KVASRIAEGF SDTALIMVDN TKFTMDCVAP TIHVYEHHEN RWRCRDPHHD Y CEDWPEAQ ...String: MPGVKLTTQA YCKMVLHGAK YPHCAVNGLL VAEKQKPRKE HLPLGGPGAH HTLFVDCIPL FHGTLALAPM LEVALTLIDS WCKDHSYVI AGYYQANERV KDASPNQVAE KVASRIAEGF SDTALIMVDN TKFTMDCVAP TIHVYEHHEN RWRCRDPHHD Y CEDWPEAQ RISASLLDSR SYETLVDFDN HLDDIRNDWT NPEINKAVLH LC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | Sample solubilized and purified in GDN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 11822 / Average exposure time: 2.66 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DARK FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
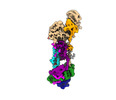
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)