[English] 日本語
 Yorodumi
Yorodumi- EMDB-40085: The Type 9 Secretion System Extended Translocon - delta GldL, Pea... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
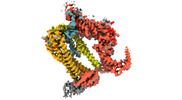
| Title | The Type 9 Secretion System Extended Translocon - delta GldL, Peak I, SprE-SkpA-Nterm SprA focused volume | |||||||||||||||||||||
 Map data Map data | volume | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | T9SS / type IX secretion system translocon / protein secretion Complex / MEMBRANE PROTEIN | |||||||||||||||||||||
| Biological species |  Flavobacterium johnsoniae UW101 (bacteria) / Flavobacterium johnsoniae UW101 (bacteria) /  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||||||||||||||
 Authors Authors | Deme JC / Lea SM | |||||||||||||||||||||
| Funding support |  United Kingdom, European Union, United Kingdom, European Union,  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2024 Journal: Nat Microbiol / Year: 2024Title: The Type 9 Secretion System caught in the act of transport Authors: Lauber F / Deme JC / Liu X / Kjaer A / Miller HL / Alcock F / Lea SM / Berks BC | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40085.map.gz emd_40085.map.gz | 273.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40085-v30.xml emd-40085-v30.xml emd-40085.xml emd-40085.xml | 26.4 KB 26.4 KB | Display Display |  EMDB header EMDB header |
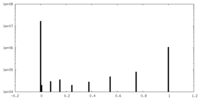
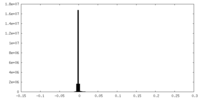
| FSC (resolution estimation) |  emd_40085_fsc.xml emd_40085_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40085.png emd_40085.png | 52.5 KB | ||
| Masks |  emd_40085_msk_1.map emd_40085_msk_1.map | 343 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40085.cif.gz emd-40085.cif.gz | 8.1 KB | ||
| Others |  emd_40085_additional_1.map.gz emd_40085_additional_1.map.gz emd_40085_additional_2.map.gz emd_40085_additional_2.map.gz emd_40085_half_map_1.map.gz emd_40085_half_map_1.map.gz emd_40085_half_map_2.map.gz emd_40085_half_map_2.map.gz | 197.3 MB 15 MB 275.2 MB 275.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40085 http://ftp.pdbj.org/pub/emdb/structures/EMD-40085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40085 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40085.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40085.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | volume | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40085_msk_1.map emd_40085_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
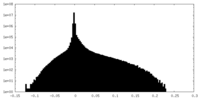
| Density Histograms |
-Additional map: local resolution filtered
| File | emd_40085_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered | ||||||||||||
| Projections & Slices |
| ||||||||||||
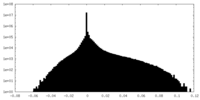
| Density Histograms |
-Additional map: post processed
| File | emd_40085_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post processed | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_40085_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_40085_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Type 9 Secretion System Extended Translocon
| Entire | Name: Type 9 Secretion System Extended Translocon |
|---|---|
| Components |
|
-Supramolecule #1: Type 9 Secretion System Extended Translocon
| Supramolecule | Name: Type 9 Secretion System Extended Translocon / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 / Details: SkpA-SprE-Nterm SprA part of complex |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae UW101 (bacteria) Flavobacterium johnsoniae UW101 (bacteria) |
-Macromolecule #1: SprE
| Macromolecule | Name: SprE / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Sequence | String: MKKNTLKYSF FLIFFLFLIA CSTKNNTFVN RNSHALSTKY NILYNGGLGL EKGLQAIKAN DQDNFWKMLP IEKMQFDENF SEGEKTKNPD FEKAETKATK AIQKHSMNIG GRERNYQIDE AYLMLGKARY YDQRFIPALE AFNYILYKYP NSSNIYTAKI WREKTNMRLG ...String: MKKNTLKYSF FLIFFLFLIA CSTKNNTFVN RNSHALSTKY NILYNGGLGL EKGLQAIKAN DQDNFWKMLP IEKMQFDENF SEGEKTKNPD FEKAETKATK AIQKHSMNIG GRERNYQIDE AYLMLGKARY YDQRFIPALE AFNYILYKYP NSSNIYTAKI WREKTNMRLG NDAIVVKNIN QLLKKTDLNK QTFSDANALL AEAFLNLEER DSAVAKLRIA EQFSRINEDR ARYKFILGQM YQEVGNKDSA TYYYDGVIHM NRKADRKYMM HAYAKKAQMY DYEKGNDTIF LKTYNKLVAD RENRPYYDVL FYEMGVFYDK KKDKENALKF YNKSLGRKSK DPYLMASAYR NIGNMYFKNT DYTMAAKYYD STLTKLNPKT REFAFIEKNR KNLDNVIKYE GIAKRNDSII KVYGMPDSER KIYFESYIAE LKKKDEAKRI LEEKEKEKLA NVERNNSASS APTAVNPNSL GKPANMDTDG IRPPSGNDAV STFYFYNPTT VAYGKLQFKK MWGNRTIGGN WRLSAIKAAN DAAMLNDSIN EAEANKLKDT VVIEKYTTAF YEKQLPKTQI AIDSIGKERN FAYYQLGLIY KEKFKEYTLA SDKLEQLLRN NPEEKLILPS MYNLYKIYQI TDPAKAEKIK SDITNNYPGS RYAQILNNTN TDDIPSPEKE YQKWYKLFQE EKFDVVLDNI DNLINQYSGD EIVSKFELLK ANTLGKVNGL EAYKKGLENV ADNYPNSDEG KNAREILEKQ VPTLERLNFT TEDNKNWKIL YLISNNDTKT LKQIEEAIRV FLLVENFERL TTSFDKYNRT QSFVAIHGLK SEAYAQDVAG VFRDDKKYKI AQPAIIISTE NYKVVQIKKN LEAYLTPKNP |
-Macromolecule #2: SkpA
| Macromolecule | Name: SkpA / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Sequence | String: TTRIGYIDME YILENVSDYK EAKSQLELKA QKWKQEIEAK KLNINSLKEG LKTEKALLTK ELIEERETEI KFQENEMLDY QQKQFGADGN LMRQKAALAK PIQDQVFTAV QDIAEAKNYD FIFDKSSDLT MLFSNKRFDI SDQVIRILNR PAMADRQKAL DERRAAREKL IE |
-Macromolecule #3: SprA
| Macromolecule | Name: SprA / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Sequence | String: MRKICIFLLV LFCGNVLRSQ VKPAVQDTTK TQFSVGKMEL ENPPSILSAY KYDPITDRYI YTTSVDGFSI DYPLVLTPKE YEDLLLKESR RDYFRKKMDA IDGKKTGAEA AKKDLLPRYY INSSLFESIF GSNTIDVKPT GSVEMDLGVR YTKQDNPAFS PRNRSSLTFD ...String: MRKICIFLLV LFCGNVLRSQ VKPAVQDTTK TQFSVGKMEL ENPPSILSAY KYDPITDRYI YTTSVDGFSI DYPLVLTPKE YEDLLLKESR RDYFRKKMDA IDGKKTGAEA AKKDLLPRYY INSSLFESIF GSNTIDVKPT GSVEMDLGVR YTKQDNPAFS PRNRSSLTFD FDQRISMSLM GKIGTRLEVN ANYDTQSTFA FQNLFKLAYT PSEDDIIQKV EVGNVSMPLN STLIRGAQSL FGVKTQLQFG RTTITGVFSE QKSQTKSVVA ENGGTVQNFD LYALDYDNDR HFFLSQYFRN KYDVSLKNYP FIDSRVQITR LEVWVTNKQN RVTTTGGGNN LRNIIALQDL GEAQVSGVPD NEVVVISSTA GFFNNPIDSP TSNTNNKYDP ATIGQAGSFL NSNIREIVTA KSGFNNTNVS EATDYSVLEN ARKLTTNEYT FNPQLGYISL QQRLANDEIL AVAFEYTVGG KVYQVGEFGS DGVDATVVTG NNSSNQAIIT QSLVLKMLKS NLTNVKNPVW NLMMKNVYQI PQAYQIKQDD FRLNILYTDP SPINYITPVQ GSSFPPNPAP DSKVEQTPLL NVFNLDRLNY NNDPQAGGDG FFDYIPGVTV DVQNGRVIFT TKEPFGELIF NKLQTGAGES YNDPTTYNAN QQKYVFRNMY RNTQAGALQD SDKNKFLLRG KYKSSGSNGI PIGAFNVPQG SVVVTAAGRV LVEGIDYSVD YQLGRVQILD PSLQASNTPI EVSLENNSIF GQQTRRFMGF NIEHKISDKF VIGGTYLKMT ERPFTQKSTY GQESVNNTIF GFNGNYSTEV PFLTRLANKL PNIDTDVPSN LSIRGEVAFL RPDAPKASDF QGEATIYVDD FEGSQSTIDM RSAYAWSLAS TPFITSINDN TFNANSNTLE YGFKRAKLSW YTIDPVFYSS KPSGISNDDL SLNTTRRIYS RELYPNTDIA QGQIQVVNTL DLTYYPGERG PYNNNPSFGA SNPSANFGGI MRALNSTNFE QGNVEYIQFW VLDPYVGNGE SPATNAGKIY FNLGEISEDV LKDGRKQYEN GLGPDQVMVN PQPLWGDVPA SQSLIYAFDT NPDNRKNQDV GLDGLPSSRE GSIYTNYAGE ADPAGDDYTY YLNADGGVLE RYKNYNGTEG NSAVSINDPN RGSTTLPDVE DINRDNTMST INAYYEYSID VKPGMQVGEN YITDIREVTN VDLPNGGTTN ARWIQFKIPV SQPQNTIGNI TDFRSIRFMR MFMTGFNSQM TVRFGALDLV RGEWRRYTGT LDANDQNPDD DGVEFDVAAV NIQENGTKCP VNYVMPPGVQ REQLYNNNTV INQNEQALAV RIGGAGLQYQ DSRAVFKNVS VDMRQYKKLK MFLHAESLPN QPTLEDDEMV GFIRFGNDFT QNFYQVEIPL KVTKTGGSCS ISPDLVWMDD NSIDLALDLL TRMKIKAMSI DINSSKRDVN GIYYPDNDPD LEGGDGDGKL TLGIKGNPNF GLVRNLMVGV KSRADHKDIK GEVWFNELRL ADLENKGGMA AILNVDTNMA DFATVSATGR KSTIGFGSLE QGANERDRED VQQYNIVTNL NLGKLLPKKW GINLPFNYAI GEEVITPEYD PFNQDIKLDQ LIRETTDQAE KDNIRTRAID YTKRKSINFI GVRKDRAPEQ KPHVYDIENF TFSQSYNQVE RHDYEVADYE DEQSNSAVNY AYTFQPKEVV PFKSTKFMKK SEYWKLLSDF NFNYLPSNIS FNTNILRQSN RQQFREVEVE GIGLDPLYRR NFAFNYQYGF GFNLTKSLKL NYSATSNNIV RNFLNDDNSP KEDFNIWDDY LDIGTPNQHA QQLVLNYDIP INKIPIFGFV KASYSYTADY MWQRSSTAFS EYEDPNGTVY DLGNTIQNSN SNTLTTTLNM NTLYKYLGLT PGAKKTAKPK TAAPPKPGEK IVNTAKPVVS SSPFYDGLIG VLTSIKNVQI NYTKNSGTVL PGYTPSVGFL GTSKPSLGFV FGSQDDVRYE AAKRGWLTTY QDFNQSFTQV SNKLLKVTAN IDLLPDLKVD LSMDRSYSEN TSEQYSVDPS TNEYKPLSPY TYGMFSISTV MIKTAFSPSD ETQSAAFDDF RSNRLIIANR LAEGHYGSGV AIPRYGDANN PIPAETDPNY AVYTANQGYP IGYTKSNQAV LLPAFLAAYT GSDASSSSTN IFRSFPIPNW SIKYNGLMRY KYFKDKFKRF SLQHNYRASY TINQFRSNFD YNSSPKVQDV NTNFYNEIIM SNVNLVEQFS PLIRMDFELK SSLRVLSEIK KDRALSMSFD NNLLTEVKGM EYIIGLGYRF KDVIFSSRLA DNPTGIIKSD INIKADFSLR NNETLVRYLD YDNNQLAAGQ NIWSLKLTAD YSFSKNLTAI FYYDHSFSKA VISTSFPLTN IRSGFTLRYN FGN |
-Macromolecule #4: PPI
| Macromolecule | Name: PPI / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Sequence | String: MKQLLTALLS LTLFISCSKD KDEVKDYTAE NEKEIVDYLA QNNLTAQRTN SGLYYIITKE GSSESEGENP GEEENTGEGE NTEENENDGH PTLNSNITVI YKGYFTNGKV FDESTEGVSY SLRTLIPGWK EGIPLLKSGG EIQLFVPAHL GYGSNGNKTV PGGAVLIFEI TLVSVN |
-Macromolecule #5: RemZ
| Macromolecule | Name: RemZ / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Sequence | String: MDVQAWGGGG AGGGASGAVL DGRAAAGGGG GAYARSNITV AAGATLNASV AGTTTNALVS GAAVNGAAGG SSTILGFETS ILALGGGGGG ANNAGGTPAG GAGGSAASSV GNVSKLDGAA GGNGVTGAIG LLTVSGAGGT AGGGGGAGGA GVASVALGNG PGNAGTAPGG ...String: MDVQAWGGGG AGGGASGAVL DGRAAAGGGG GAYARSNITV AAGATLNASV AGTTTNALVS GAAVNGAAGG SSTILGFETS ILALGGGGGG ANNAGGTPAG GAGGSAASSV GNVSKLDGAA GGNGVTGAIG LLTVSGAGGT AGGGGGAGGA GVASVALGNG PGNAGTAPGG GGSGAMQSLL GGAQIGGSGA AGRVIITYTC PTYSITGISA ANVCNSVGTT SVVTLTSSGG GLPIGPYVVT YNRSNPSGTG LTAIMNVTTP GTGTFTAAGL NVIGTSNITV TNLTSAACSS NISTNNVASL TVFAATVGGT LAGTATVCSG ATSGTLTLSG QTGSIIKWES SVSPFTVWTT IPNTTNTYTS GALTETSQFR AVIQNGNCAV VNSSIATITV NPLPQGSLSA NGPFCVTGSG QLTFTATAGT GPYTIVYKEN GGADRTAANI SSGVAFPTFT TPVTTTTVYT LVSVTGANTC SRSSGFTNNT ATITVNSRIA TPGFGTVTQP DCVTSTGSVV LTGLPAGSWT ITQSGTASQT YNSSGTTYTI SNLAVGNYTF TVQDAANCPS LATSTLTLIA PVVNIWNGTS WSKGSPPIST DVVRFSGNYS TTGNLSGCSL IVDSGFTVTV NSNHTLTISN AVTNNGGQLI FENNSSLLQT NNVTNVGNIT YKRITPPVRR YDLTYWSSPI TRTPPFTLYD LSPGTLADKY YSYDPVAGWV ISFNGTQQMV PGRGYVVRAP QTNDLNTGAN YLGAFVGVPN NGPISVSLGT AEAFQLLGNP YPSAIYADQF IANNSANLYG TLYFWTHNSL PSSSTPGGAQ YNYDNNDYAV YNLSGSIIVG GMTGQGATTP GNQSAPLGYI AAGQGFFVVS KTAGNAVFTN SMRVAANNTQ FYKTNKSAIE RHRVWINLTN TQGAFKQLLI GYIEGATNFW DHNYDAITAD ANPHLDFYSI NEGQNLVIQG RSLPFNESDV VPLGYRSAIA GEFSISLDHA DGDLTNHAVY LEDKLTNTLH NLQTSNYTFN TAIGTFSDRF VIRYTTATLG TDDFENQTNS FYVSVKDKTI KLNSTEDVMR EVSIFDISGK LLYNNKKVEN TEFQVSNFQS GNQVLIVKVT LDNGNIITKK IVFN |
-Macromolecule #6: PorV
| Macromolecule | Name: PorV / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flavobacterium johnsoniae (bacteria) Flavobacterium johnsoniae (bacteria) |
| Sequence | String: MKKISLLLIC LLITTFAKAQ DIERPITTGV PFLLVAADAR AAGLGDQGVA TSSDVFSQQW NPAKYAFAED AQGLSISYTP YLTDLANDIS LGQVTYYNKI NDRSAFAGSF RYFGFGGIEL RQTGDPNEPT REVNPNEFAL DGSYSLKLSE TFSMAVAARY IRSNLKVATE ...String: MKKISLLLIC LLITTFAKAQ DIERPITTGV PFLLVAADAR AAGLGDQGVA TSSDVFSQQW NPAKYAFAED AQGLSISYTP YLTDLANDIS LGQVTYYNKI NDRSAFAGSF RYFGFGGIEL RQTGDPNEPT REVNPNEFAL DGSYSLKLSE TFSMAVAARY IRSNLKVATE EIDASAAGSF AVDVAGFYQS EEIAYSDFNG RWRAGFNIQN LGPKISYDHD DLSANFLPAN LRVGGGFDFI FDDYNKLGVS LELTKLLVPT PPGPGTPYDA NGDGDFTDPG DISQSQADEA NYKKYKDIGW VSGIFKSFGD APGGFSEELK EITYSAAAEY MYQDAFAMRL GYYHESPMKG AKQFFSLGAG FKYSMIKVDV SYLFSASKVK NPLENTLRFS LTFNFGDKYE TY |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 61.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)