[English] 日本語
 Yorodumi
Yorodumi- EMDB-39858: Cryo-EM structure of the insect olfactory receptor OR5-Orco heter... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MEMBRANE PROTEIN | |||||||||
| Function / homology | Olfactory receptor, insect / 7tm Odorant receptor / olfactory receptor activity / odorant binding / signal transduction / plasma membrane / Odorant receptor / Odorant receptor Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Wang YD / Qiu L / Guan ZY / Wang Q / Wang GR / Yin P | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex. Authors: Yidong Wang / Liang Qiu / Bing Wang / Zeyuan Guan / Zhi Dong / Jie Zhang / Song Cao / Lulu Yang / Bo Wang / Zhou Gong / Liwei Zhang / Weihua Ma / Zhu Liu / Delin Zhang / Guirong Wang / Ping Yin /  Abstract: Insects detect and discriminate a diverse array of chemicals using odorant receptors (ORs), which are ligand-gated ion channels comprising a divergent odorant-sensing OR and a conserved odorant ...Insects detect and discriminate a diverse array of chemicals using odorant receptors (ORs), which are ligand-gated ion channels comprising a divergent odorant-sensing OR and a conserved odorant receptor co-receptor (Orco). In this work, we report structures of the OR5-Orco heterocomplex from the pea aphid alone and bound to its known activating ligand, geranyl acetate. In these structures, three Orco subunits serve as scaffold components that cannot bind the ligand and remain relatively unchanged. Upon ligand binding, the pore-forming helix S7b of OR5 shifts outward from the central pore axis, causing an asymmetrical pore opening for ion influx. Our study provides insights into odorant recognition and channel gating of the OR-Orco heterocomplex and offers structural resources to support development of innovative insecticides and repellents for pest control. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39858.map.gz emd_39858.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39858-v30.xml emd-39858-v30.xml emd-39858.xml emd-39858.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39858.png emd_39858.png | 45.1 KB | ||
| Filedesc metadata |  emd-39858.cif.gz emd-39858.cif.gz | 5.9 KB | ||
| Others |  emd_39858_half_map_1.map.gz emd_39858_half_map_1.map.gz emd_39858_half_map_2.map.gz emd_39858_half_map_2.map.gz | 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39858 http://ftp.pdbj.org/pub/emdb/structures/EMD-39858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39858 | HTTPS FTP |
-Related structure data
| Related structure data |  8z9aMC  8z9zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39858.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39858.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
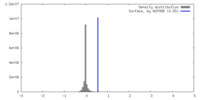
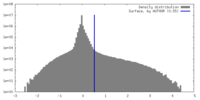
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_39858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
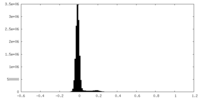
| Density Histograms |
-Half map: #2
| File | emd_39858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
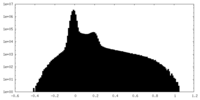
| Density Histograms |
- Sample components
Sample components
-Entire : OR-Orco heterocomplex
| Entire | Name: OR-Orco heterocomplex |
|---|---|
| Components |
|
-Supramolecule #1: OR-Orco heterocomplex
| Supramolecule | Name: OR-Orco heterocomplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Odorant receptor, ApisOrco
| Macromolecule | Name: Odorant receptor, ApisOrco / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.916766 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGYKKDGLIK DLWPNIRLIQ LSGLFISEYY DDYSGLAVLF RKIYSWITAI IIYSQFIFIV IFMVTKSNDS DQLAAGVVTT LFFTHSMIK FVYFSTGTKS FYRTLSCWNN TSPHPLFAES HSRFHAKSLS RMRQLLIIVS IVTIFTTISW TTITFFGESV W KVPDPETF ...String: MGYKKDGLIK DLWPNIRLIQ LSGLFISEYY DDYSGLAVLF RKIYSWITAI IIYSQFIFIV IFMVTKSNDS DQLAAGVVTT LFFTHSMIK FVYFSTGTKS FYRTLSCWNN TSPHPLFAES HSRFHAKSLS RMRQLLIIVS IVTIFTTISW TTITFFGESV W KVPDPETF NQTMYVPVPR LMLHSWYPWD SGHGLGYIVA FVLQFYWVFI TLSHSNLMEL LFSSFLVHAC EQLQHLKEIL NP LIELSAT LDSSVHNPAE IFRANSAKNQ SINGIDHDYN GSYVNEITEY GTKGENEPNR KGPNNLTSNQ EVLVRSAIKY WVE RHKHVV KYVSLITECY GSALLFHMLV STVILTILAY QATKINGVNV FAFSTIGYLM YSFAQIFMFC IHGNELIEES SSVM EAAYG CHWYDGSEEA KTFVQIVCQQ CQKPLIVSGA KFFNVSLDLF ASVLGAVVTY FMVLVQLK UniProtKB: Odorant receptor |
-Macromolecule #2: Odorant receptor, ApisOR5
| Macromolecule | Name: Odorant receptor, ApisOR5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.45407 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQRIDTINMF LQMTGCTDSK AMLYLTYFEF LITFYYLIAT YASIVHFEQS VTIQLFALLC MLIECVILLN ITFRLYHKNH IREMHQYSR RLGIPDSYRS VINVITKYHL IASNIFVVFP VTYAIFCDSV RVGDPFTFPF LDVLPMHTDN LAIYACKYLV Y AISVYIAH ...String: MQRIDTINMF LQMTGCTDSK AMLYLTYFEF LITFYYLIAT YASIVHFEQS VTIQLFALLC MLIECVILLN ITFRLYHKNH IREMHQYSR RLGIPDSYRS VINVITKYHL IASNIFVVFP VTYAIFCDSV RVGDPFTFPF LDVLPMHTDN LAIYACKYLV Y AISVYIAH VELCFINTTF IYYVGVLKHR LETIVQTIGE AFADNDEQKF KYAIIQHQKL LSYFNTMKIV FSKPILLSMS FN AIYFGLT TSFVIQAIRG YINQAILSIC IASSAAAVIN ITIYTFYGSE LMDLHDKILH VLFDNAFFYV SKSFKSSILI MMT RVTIPL KFTVGYIFTI NLNLLLKILK MSYTVLNVLL SSETIKPHKL S UniProtKB: Odorant receptor |
-Macromolecule #3: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 3 / Number of copies: 7 / Formula: PC1 |
|---|---|
| Molecular weight | Theoretical: 790.145 Da |
| Chemical component information |  ChemComp-PC1: |
-Macromolecule #4: [(2E)-3,7-dimethylocta-2,6-dienyl] ethanoate
| Macromolecule | Name: [(2E)-3,7-dimethylocta-2,6-dienyl] ethanoate / type: ligand / ID: 4 / Number of copies: 1 / Formula: SOU |
|---|---|
| Molecular weight | Theoretical: 196.286 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 399858 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)