+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of APJR complex with agonistic antibody | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / APJ / Agonistic Antibody / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information: / apelin receptor activity / apelin receptor signaling pathway / mechanoreceptor activity / regulation of gap junction assembly / positive regulation of G protein-coupled receptor internalization / vascular associated smooth muscle cell differentiation / atrioventricular valve development / regulation of body fluid levels / venous blood vessel development ...: / apelin receptor activity / apelin receptor signaling pathway / mechanoreceptor activity / regulation of gap junction assembly / positive regulation of G protein-coupled receptor internalization / vascular associated smooth muscle cell differentiation / atrioventricular valve development / regulation of body fluid levels / venous blood vessel development / positive regulation of cardiac muscle hypertrophy in response to stress / endocardial cushion formation / positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis / coronary vasculature development / adult heart development / vasculature development / G protein-coupled peptide receptor activity / aorta development / negative regulation of cardiac muscle hypertrophy in response to stress / ventricular septum morphogenesis / blood vessel development / heart looping / vasculogenesis / gastrulation / Peptide ligand-binding receptors / positive regulation of release of sequestered calcium ion into cytosol / electron transport chain / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / positive regulation of angiogenesis / signaling receptor activity / heart development / regulation of gene expression / angiogenesis / G alpha (i) signalling events / electron transfer activity / periplasmic space / iron ion binding / G protein-coupled receptor signaling pathway / negative regulation of gene expression / heme binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.01 Å | |||||||||
 Authors Authors | Yue Y / Liu LE / Wu LJ / Xu F | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structural insights into the regulation of monomeric and dimeric apelin receptor. Authors: Yang Yue / Lier Liu / Lijie Wu / Chanjuan Xu / Man Na / Shenhui Liu / Yuxuan Liu / Fei Li / Junlin Liu / Songting Shi / Hui Lei / Minxuan Zhao / Tianjie Yang / Wei Ji / Arthur Wang / Michael ...Authors: Yang Yue / Lier Liu / Lijie Wu / Chanjuan Xu / Man Na / Shenhui Liu / Yuxuan Liu / Fei Li / Junlin Liu / Songting Shi / Hui Lei / Minxuan Zhao / Tianjie Yang / Wei Ji / Arthur Wang / Michael A Hanson / Raymond C Stevens / Jianfeng Liu / Fei Xu /   Abstract: The apelin receptor (APJR) emerges as a promising drug target for cardiovascular health and muscle regeneration. While prior research unveiled the structural versatility of APJR in coupling to Gi ...The apelin receptor (APJR) emerges as a promising drug target for cardiovascular health and muscle regeneration. While prior research unveiled the structural versatility of APJR in coupling to Gi proteins as a monomer or dimer, the dynamic regulation within the APJR dimer during activation remains poorly understood. In this study, we present the structures of the APJR dimer and monomer complexed with its endogenous ligand apelin-13. In the dimeric structure, apelin-13 binds exclusively to one protomer that is coupled with Gi proteins, revealing a distinct ligand-binding behavior within APJR homodimers. Furthermore, binding of an antagonistic antibody induces a more compact dimerization by engaging both protomers. Notably, structural analyses of the APJR dimer complexed with an agonistic antibody, with or without Gi proteins, suggest that G protein coupling may promote the dissociation of the APJR dimer during activation. These findings underscore the intricate interplay between ligands, dimerization, and G protein coupling in regulating APJR signaling pathways. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39810.map.gz emd_39810.map.gz | 56.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39810-v30.xml emd-39810-v30.xml emd-39810.xml emd-39810.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39810_fsc.xml emd_39810_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_39810.png emd_39810.png | 110.5 KB | ||
| Filedesc metadata |  emd-39810.cif.gz emd-39810.cif.gz | 6.1 KB | ||
| Others |  emd_39810_half_map_1.map.gz emd_39810_half_map_1.map.gz emd_39810_half_map_2.map.gz emd_39810_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39810 http://ftp.pdbj.org/pub/emdb/structures/EMD-39810 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39810 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39810 | HTTPS FTP |
-Related structure data
| Related structure data |  8z74MC  8xqeC  8xqfC  8xqiC  8xqjC  8z7jC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39810.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39810.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_39810_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
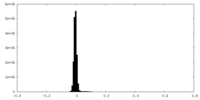
| Density Histograms |
-Half map: #2
| File | emd_39810_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
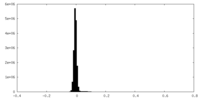
| Density Histograms |
- Sample components
Sample components
-Entire : APJR complex with antibody
| Entire | Name: APJR complex with antibody |
|---|---|
| Components |
|
-Supramolecule #1: APJR complex with antibody
| Supramolecule | Name: APJR complex with antibody / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: agonistic antibody
| Macromolecule | Name: agonistic antibody / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.929495 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: QVQLVESGGG SVQSGGSLTL SCAASGSTYS SHCMGWFRQA PGKEREGVAL MTRSRGTSYA DSVKGRFTIS QDNTKNILYL QMNSLKPED TAMYYCAAVP RAGIEYSGAY CKWNMKDSGS WGQGTQVTVS S |
-Macromolecule #2: Soluble cytochrome b562,Apelin receptor
| Macromolecule | Name: Soluble cytochrome b562,Apelin receptor / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.312578 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHLEVLFQ GPADLEDNWE TLNDNLKVIE KADNAAQVKD ALTKMRAAAL DAQKATPPK LEDKSPDSPE MKDFRHGFDI LVGQIDDALK LANEGKVKEA QAAAEQLKTT RNAYIQKYLE EGGDFDNYYG A DNQSECEY ...String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHLEVLFQ GPADLEDNWE TLNDNLKVIE KADNAAQVKD ALTKMRAAAL DAQKATPPK LEDKSPDSPE MKDFRHGFDI LVGQIDDALK LANEGKVKEA QAAAEQLKTT RNAYIQKYLE EGGDFDNYYG A DNQSECEY TDWKSSGALI PAIYMLVFLL GTTGNGLVLW TVFRSSREKR RSADIFIASL AVADLTFVVT LPLWATYTYR DY DWPFGTF FCKLSSYLIF VNMYASVFCL TGLSFDRYLA IVRPVANARL RLRVSGAVAT AVLWVLAALL AMPVMVLRTT GDL ENTTKV QCYMDYSMVA TVSSEWAWEV GLGVSSTTVG FVVPFTIMLT CYFFIAQTIA GHFRKERIEG LRKRRRLLSI IVVL VVTFA LCWMPYHLVK TLYMLGSLLH WPCDFDLFLM NIFPYCTCIS YVNSCLNPFL YAFFDPRFRQ ACTSMLCCGQ SR UniProtKB: Soluble cytochrome b562, Apelin receptor |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)