+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human phosphorylase kinase - inactive state | |||||||||
 Map data Map data | EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinase / glycogenolysis / SIGNALING PROTEIN / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphorylase kinase / phosphorylase kinase activity / phosphorylase kinase complex / transporter inhibitor activity / positive regulation of glycogen catabolic process / tau-protein kinase / : / type 3 metabotropic glutamate receptor binding / response to corticosterone / negative regulation of high voltage-gated calcium channel activity ...phosphorylase kinase / phosphorylase kinase activity / phosphorylase kinase complex / transporter inhibitor activity / positive regulation of glycogen catabolic process / tau-protein kinase / : / type 3 metabotropic glutamate receptor binding / response to corticosterone / negative regulation of high voltage-gated calcium channel activity / glycogen metabolic process / Glycogen breakdown (glycogenolysis) / negative regulation of calcium ion export across plasma membrane / regulation of cardiac muscle cell action potential / nitric-oxide synthase binding / presynaptic endocytosis / regulation of synaptic vesicle exocytosis / regulation of cell communication by electrical coupling involved in cardiac conduction / calcineurin-mediated signaling / adenylate cyclase binding / protein phosphatase activator activity / regulation of synaptic vesicle endocytosis / detection of calcium ion / regulation of cardiac muscle contraction / postsynaptic cytosol / catalytic complex / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / phosphatidylinositol 3-kinase binding / presynaptic cytosol / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / titin binding / regulation of calcium-mediated signaling / voltage-gated potassium channel complex / sperm midpiece / calcium channel complex / substantia nigra development / regulation of heart rate / response to amphetamine / calyx of Held / adenylate cyclase activator activity / nitric-oxide synthase regulator activity / sarcomere / protein serine/threonine kinase activator activity / regulation of cytokinesis / spindle microtubule / generation of precursor metabolites and energy / calcium channel regulator activity / response to calcium ion / mitochondrial membrane / Schaffer collateral - CA1 synapse / G2/M transition of mitotic cell cycle / long-term synaptic potentiation / spindle pole / calcium-dependent protein binding / synaptic vesicle membrane / myelin sheath / growth cone / vesicle / carbohydrate metabolic process / transmembrane transporter binding / calmodulin binding / non-specific serine/threonine protein kinase / G protein-coupled receptor signaling pathway / protein domain specific binding / protein serine kinase activity / calcium ion binding / centrosome / protein kinase binding / enzyme binding / signal transduction / protein-containing complex / ATP binding / nucleus / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Ma R / Yan K | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Molecular basis for the regulation of human phosphorylase kinase by phosphorylation and Ca. Authors: Ruifang Ma / Bowen Du / Chen Shi / Lei Wang / Fuxing Zeng / Jie Han / Huiyi Guan / Yong Wang / Kaige Yan /  Abstract: Phosphorylase kinase (PhK) regulates the degradation of glycogen by integrating diverse signals, providing energy to the organism. Dysfunctional mutations may directly lead to Glycogen Storage ...Phosphorylase kinase (PhK) regulates the degradation of glycogen by integrating diverse signals, providing energy to the organism. Dysfunctional mutations may directly lead to Glycogen Storage Disease type IX (GSD IX), whereas the abnormal expression of PhK is also associated with tumors. Here, we use cryo-electron microscopy (cryo-EM) to resolve its near-atomic structures in the inactive and active states. These structures reveal the interactions and relative locations of the four subunits (αβγδ) within the PhK complex. Phosphorylated α and β subunits induce PhK to present a more compact state, while Ca causes sliding of the δ subunit along the helix of the γ subunit. Both actions synergistically activate PhK by enabling the de-inhibition of the γ subunit. We also identified different binding modes between PhK and its substrate, glycogen phosphorylase (GP), in two distinct states, using cross-linking mass spectrometry (XL-MS). This study provides valuable insights into the regulatory mechanisms of PhK, thereby enhancing our understanding of GSD IX and its implications in tumorigenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39777.map.gz emd_39777.map.gz | 398.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39777-v30.xml emd-39777-v30.xml emd-39777.xml emd-39777.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39777_fsc.xml emd_39777_fsc.xml | 15.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_39777.png emd_39777.png | 27.4 KB | ||
| Masks |  emd_39777_msk_1.map emd_39777_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-39777.cif.gz emd-39777.cif.gz | 7.9 KB | ||
| Others |  emd_39777_half_map_1.map.gz emd_39777_half_map_1.map.gz emd_39777_half_map_2.map.gz emd_39777_half_map_2.map.gz | 391.3 MB 391.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39777 http://ftp.pdbj.org/pub/emdb/structures/EMD-39777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39777 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39777 | HTTPS FTP |
-Validation report
| Summary document |  emd_39777_validation.pdf.gz emd_39777_validation.pdf.gz | 934.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39777_full_validation.pdf.gz emd_39777_full_validation.pdf.gz | 933.8 KB | Display | |
| Data in XML |  emd_39777_validation.xml.gz emd_39777_validation.xml.gz | 25.3 KB | Display | |
| Data in CIF |  emd_39777_validation.cif.gz emd_39777_validation.cif.gz | 33.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39777 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39777 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39777 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39777 | HTTPS FTP |
-Related structure data
| Related structure data |  8z5pMC  8z5mC  8z5qC  8z5rC  8z5tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39777.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39777.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
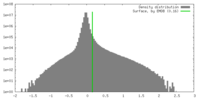
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_39777_msk_1.map emd_39777_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
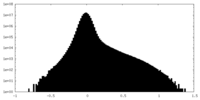
| Density Histograms |
-Half map: half map
| File | emd_39777_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_39777_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human phosphorylase kinase - inactive state
| Entire | Name: human phosphorylase kinase - inactive state |
|---|---|
| Components |
|
-Supramolecule #1: human phosphorylase kinase - inactive state
| Supramolecule | Name: human phosphorylase kinase - inactive state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.3 MDa |
-Macromolecule #1: Phosphorylase b kinase regulatory subunit alpha, skeletal muscle ...
| Macromolecule | Name: Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 137.469422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRSRSNSGVR LDGYARLVQQ TILCHQNPVT GLLPASYDQK DAWVRDNVYS ILAVWGLGLA YRKNADRDED KAKAYELEQS VVKLMRGLL HCMIRQVDKV ESFKYSQSTK DSLHAKYNTK TCATVVGDDQ WGHLQLDATS VYLLFLAQMT ASGLHIIHSL D EVNFIQNL ...String: MRSRSNSGVR LDGYARLVQQ TILCHQNPVT GLLPASYDQK DAWVRDNVYS ILAVWGLGLA YRKNADRDED KAKAYELEQS VVKLMRGLL HCMIRQVDKV ESFKYSQSTK DSLHAKYNTK TCATVVGDDQ WGHLQLDATS VYLLFLAQMT ASGLHIIHSL D EVNFIQNL VFYIEAAYKT ADFGIWERGD KTNQGISELN ASSVGMAKAA LEALDELDLF GVKGGPQSVI HVLADEVQHC QS ILNSLLP RASTSKEVDA SLLSVVSFPA FAVEDSQLVE LTKQEIITKL QGRYGCCRFL RDGYKTPKED PNRLYYEPAE LKL FENIEC EWPLFWTYFI LDGVFSGNAE QVQEYKEALE AVLIKGKNGV PLLPELYSVP PDRVDEEYQN PHTVDRVPMG KLPH MWGQS LYILGSLMAE GFLAPGEIDP LNRRFSTVPK PDVVVQVSIL AETEEIKTIL KDKGIYVETI AEVYPIRVQP ARILS HIYS SLGCNNRMKL SGRPYRHMGV LGTSKLYDIR KTIFTFTPQF IDQQQFYLAL DNKMIVEMLR TDLSYLCSRW RMTGQP TIT FPISHSMLDE DGTSLNSSIL AALRKMQDGY FGGARVQTGK LSEFLTTSCC THLSFMDPGP EGKLYSEDYD DNYDYLE SG NWMNDYDSTS HARCGDEVAR YLDHLLAHTA PHPKLAPTSQ KGGLDRFQAA VQTTCDLMSL VTKAKELHVQ NVHMYLPT K LFQASRPSFN LLDSPHPRQE NQVPSVRVEI HLPRDQSGEV DFKALVLQLK ETSSLQEQAD ILYMLYTMKG PDWNTELYN ERSATVRELL TELYGKVGEI RHWGLIRYIS GILRKKVEAL DEACTDLLSH QKHLTVGLPP EPREKTISAP LPYEALTQLI DEASEGDMS ISILTQEIMV YLAMYMRTQP GLFAEMFRLR IGLIIQVMAT ELAHSLRCSA EEATEGLMNL SPSAMKNLLH H ILSGKEFG VERSVRPTDS NVSPAISIHE IGAVGATKTE RTGIMQLKSE IKQVEFRRLS ISAESQSPGT SMTPSSGSFP SA YDQQSSK DSRQGQWQRR RRLDGALNRV PVGFYQKVWK VLQKCHGLSV EGFVLPSSTT REMTPGEIKF SVHVESVLNR VPQ PEYRQL LVEAILVLTM LADIEIHSIG SIIAVEKIVH IANDLFLQEQ KTLGADDTML AKDPASGICT LLYDSAPSGR FGTM TYLSK AAATYVQEFL PHSICAMQ UniProtKB: Phosphorylase b kinase regulatory subunit alpha, skeletal muscle isoform |
-Macromolecule #2: Phosphorylase b kinase regulatory subunit beta
| Macromolecule | Name: Phosphorylase b kinase regulatory subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 125.032961 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGAAGLTAE VSWKVLERRA RTKRSGSVYE PLKSINLPRP DNETLWDKLD HYYRIVKSTL LLYQSPTTGL FPTKTCGGDQ KAKIQDSLY CAAGAWALAL AYRRIDDDKG RTHELEHSAI KCMRGILYCY MRQADKVQQF KQDPRPTTCL HSVFNVHTGD E LLSYEEYG ...String: MAGAAGLTAE VSWKVLERRA RTKRSGSVYE PLKSINLPRP DNETLWDKLD HYYRIVKSTL LLYQSPTTGL FPTKTCGGDQ KAKIQDSLY CAAGAWALAL AYRRIDDDKG RTHELEHSAI KCMRGILYCY MRQADKVQQF KQDPRPTTCL HSVFNVHTGD E LLSYEEYG HLQINAVSLY LLYLVEMISS GLQIIYNTDE VSFIQNLVFC VERVYRVPDF GVWERGSKYN NGSTELHSSS VG LAKAALE AINGFNLFGN QGCSWSVIFV DLDAHNRNRQ TLCSLLPRES RSHNTDAALL PCISYPAFAL DDEVLFSQTL DKV VRKLKG KYGFKRFLRD GYRTSLEDPN RCYYKPAEIK LFDGIECEFP IFFLYMMIDG VFRGNPKQVQ EYQDLLTPVL HHTT EGYPV VPKYYYVPAD FVEYEKNNPG SQKRFPSNCG RDGKLFLWGQ ALYIIAKLLA DELISPKDID PVQRYVPLKD QRNVS MRFS NQGPLENDLV VHVALIAESQ RLQVFLNTYG IQTQTPQQVE PIQIWPQQEL VKAYLQLGIN EKLGLSGRPD RPIGCL GTS KIYRILGKTV VCYPIIFDLS DFYMSQDVFL LIDDIKNALQ FIKQYWKMHG RPLFLVLIRE DNIRGSRFNP ILDMLAA LK KGIIGGVKVH VDRLQTLISG AVVEQLDFLR ISDTEELPEF KSFEELEPPK HSKVKRQSST PSAPELGQQP DVNISEWK D KPTHEILQKL NDCSCLASQA ILLGILLKRE GPNFITKEGT VSDHIERVYR RAGSQKLWLA VRYGAAFTQK FSSSIAPHI TTFLVHGKQV TLGAFGHEEE VISNPLSPRV IQNIIYYKCN THDEREAVIQ QELVIHIGWI ISNNPELFSG MLKIRIGWII HAMEYELQI RGGDKPALDL YQLSPSEVKQ LLLDILQPQQ NGRCWLNRRQ IDGSLNRTPT GFYDRVWQIL ERTPNGIIVA G KHLPQQPT LSDMTMYEMN FSLLVEDTLG NIDQPQYRQI VVELLMVVSI VLERNPELEF QDKVDLDRLV KEAFNEFQKD QS RLKEIEK QDDMTSFYNT PPLGKRGTCS YLTKAVMNLL LEGEVKPNND DPCLIS UniProtKB: Phosphorylase b kinase regulatory subunit beta |
-Macromolecule #3: Phosphorylase b kinase gamma catalytic chain, skeletal muscle/hea...
| Macromolecule | Name: Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: phosphorylase kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.084672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTRDEALPDS HSAQDFYENY EPKEILGRGV SSVVRRCIHK PTSQEYAVKV IDVTGGGSFS PEEVRELREA TLKEVDILRK VSGHPNIIQ LKDTYETNTF FFLVFDLMKR GELFDYLTEK VTLSEKETRK IMRALLEVIC TLHKLNIVHR DLKPENILLD D NMNIKLTD ...String: MTRDEALPDS HSAQDFYENY EPKEILGRGV SSVVRRCIHK PTSQEYAVKV IDVTGGGSFS PEEVRELREA TLKEVDILRK VSGHPNIIQ LKDTYETNTF FFLVFDLMKR GELFDYLTEK VTLSEKETRK IMRALLEVIC TLHKLNIVHR DLKPENILLD D NMNIKLTD FGFSCQLEPG ERLREVCGTP SYLAPEIIEC SMNEDHPGYG KEVDMWSTGV IMYTLLAGSP PFWHRKQMLM LR MIMSGNY QFGSPEWDDY SDTVKDLVSR FLVVQPQNRY TAEEALAHPF FQQYLVEEVR HFSPRGKFKV IALTVLASVR IYY QYRRVK PVTREIVIRD PYALRPLRRL IDAYAFRIYG HWVKKGQQQN RAALFENTPK AVLLSLAEED Y UniProtKB: Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform |
-Macromolecule #4: Calmodulin-3
| Macromolecule | Name: Calmodulin-3 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.921584 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDKSG PDEVDASGRM ADQLTEEQIA EFKEAFSLFD KDGDGTITTK ELGTVMRSLG QNPTEAELQD MINEVDADGN GTIDFPEFL TMMARKMKDT DSEEEIREAF RVFDKDGNGY ISAAELRHVM TNLGEKLTDE EVDEMIREAD IDGDGQVNYE E FVQMMTAK UniProtKB: Calmodulin-3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)