[English] 日本語
 Yorodumi
Yorodumi- EMDB-39773: Cryo-EM structure of E.coli SPFH-NfeD family protein complex QmcA-YbbJ -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of E.coli SPFH-NfeD family protein complex QmcA-YbbJ | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Qiao Z / Gao YG | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2024 Journal: Structure / Year: 2024Title: Cryo-EM structure of the SPFH-NfeD family protein complex QmcA-YbbJ. Authors: Kwan Ann Tan / Zhu Qiao / Zachary Ze En Lim / Joshua Yi Yeo / Yonlada Yong / Phong Hoa Do / Rya Ero / Yong-Gui Gao /  Abstract: The SPFH (stomatin, prohibitin, flotillin, and HflK/C) protein family is universally present and encompasses the evolutionarily conserved SPFH domain. These proteins are predominantly localized in ...The SPFH (stomatin, prohibitin, flotillin, and HflK/C) protein family is universally present and encompasses the evolutionarily conserved SPFH domain. These proteins are predominantly localized in lipid raft and implicated in various biological processes. The NfeD (nodulation formation efficiency D) protein family is often encoded in tandem with SPFH proteins, suggesting a close functional relationship. Here, we elucidate the cryoelectron microscopy (cryo-EM) structure of the Escherichia coli QmcA-YbbJ complex belonging to the SPFH and NfeD families, respectively. Our findings reveal that the QmcA-YbbJ complex forms an intricate cage-like structure composed of 26 copies of QmcA-YbbJ heterodimers. The transmembrane helices of YbbJ act as adhesive elements bridging adjacent QmcA molecules, while the oligosaccharide-binding domain of YbbJ encapsulates the SPFH domain of QmcA. Our structural study significantly contributes to understanding the functional role of the NfeD protein family and sheds light on the interplay between SPFH and NfeD family proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39773.map.gz emd_39773.map.gz | 57.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39773-v30.xml emd-39773-v30.xml emd-39773.xml emd-39773.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39773.png emd_39773.png | 94.2 KB | ||
| Filedesc metadata |  emd-39773.cif.gz emd-39773.cif.gz | 5.4 KB | ||
| Others |  emd_39773_half_map_1.map.gz emd_39773_half_map_1.map.gz emd_39773_half_map_2.map.gz emd_39773_half_map_2.map.gz | 48.3 MB 48.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39773 http://ftp.pdbj.org/pub/emdb/structures/EMD-39773 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39773 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39773 | HTTPS FTP |
-Validation report
| Summary document |  emd_39773_validation.pdf.gz emd_39773_validation.pdf.gz | 858.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39773_full_validation.pdf.gz emd_39773_full_validation.pdf.gz | 858.3 KB | Display | |
| Data in XML |  emd_39773_validation.xml.gz emd_39773_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_39773_validation.cif.gz emd_39773_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39773 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39773 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39773 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39773 | HTTPS FTP |
-Related structure data
| Related structure data |  8z5gMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39773.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39773.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.46 Å | ||||||||||||||||||||||||||||||||||||
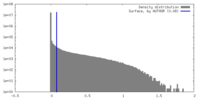
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39773_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
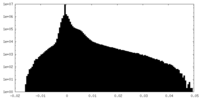
| Density Histograms |
-Half map: #1
| File | emd_39773_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : QmcA-YbbJ complex
| Entire | Name: QmcA-YbbJ complex |
|---|---|
| Components |
|
-Supramolecule #1: QmcA-YbbJ complex
| Supramolecule | Name: QmcA-YbbJ complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein QmcA
| Macromolecule | Name: Protein QmcA / type: protein_or_peptide / ID: 1 / Number of copies: 26 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.778023 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLIFIPILIF VALVIVGAGV KIVPQGYQWT VERFGRYTKT LQPGLSLVVP FMDRIGRKIN MMEQVLDIPS QEVISKDNAN VTIDAVCFI QVIDAPRAAY EVSNLELAII NLTMTNIRTV LGSMELDEML SQRDSINSRL LRIVDEATNP WGIKVTRIEI R DVRPPAEL ...String: MLIFIPILIF VALVIVGAGV KIVPQGYQWT VERFGRYTKT LQPGLSLVVP FMDRIGRKIN MMEQVLDIPS QEVISKDNAN VTIDAVCFI QVIDAPRAAY EVSNLELAII NLTMTNIRTV LGSMELDEML SQRDSINSRL LRIVDEATNP WGIKVTRIEI R DVRPPAEL ISSMNAQMKA ERTKRAYILE AEGIRQAEIL KAEGEKQSQI LKAEGERQSA FLQAEARERS AEAEARATKM VS EAIASGD IQAVNYFVAQ KYTEALQQIG SSSNSKVVMM PLEASSLMGS IAGIAELVKD SANKRTQP UniProtKB: Protein QmcA |
-Macromolecule #2: Inner membrane protein YbbJ
| Macromolecule | Name: Inner membrane protein YbbJ / type: protein_or_peptide / ID: 2 / Number of copies: 26 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.956775 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMELMVVHPH IFWLSLGGLL LAAEMLGGNG YLLWSGVAAV ITGLVVWLVP LGWEWQGVMF AILTLLAAWL WWKWLSRRVR EQKHSDSHL NQRGQQLIGR RFVLESPLVN GRGHMRVGDS SWPVSASEDL GAGTHVEVIA IEGITLHIRA VSS UniProtKB: Inner membrane protein YbbJ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 36372 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)