[English] 日本語
 Yorodumi
Yorodumi- EMDB-39703: Cryo-EM structure of ATP-bound human very long-chain fatty acid A... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ATP-bound human very long-chain fatty acid ABC transporter ABCD3 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | very long-chain fatty / Peroxisome / ABC transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphytanic acid metabolic process / long-chain fatty acid import into peroxisome / very long-chain fatty acid catabolic process / very long-chain fatty acid metabolic process / Class I peroxisomal membrane protein import / peroxisome organization / fatty acyl-CoA hydrolase activity / ABC transporters in lipid homeostasis / bile acid biosynthetic process / Hydrolases; Acting on ester bonds; Thioester hydrolases ...phytanic acid metabolic process / long-chain fatty acid import into peroxisome / very long-chain fatty acid catabolic process / very long-chain fatty acid metabolic process / Class I peroxisomal membrane protein import / peroxisome organization / fatty acyl-CoA hydrolase activity / ABC transporters in lipid homeostasis / bile acid biosynthetic process / Hydrolases; Acting on ester bonds; Thioester hydrolases / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / peroxisomal membrane / long-chain fatty acid transmembrane transporter activity / bile acid and bile salt transport / fatty acid beta-oxidation / RHOC GTPase cycle / peroxisomal matrix / RHOA GTPase cycle / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / fatty acid biosynthetic process / peroxisome / response to xenobiotic stimulus / intracellular membrane-bounded organelle / protein homodimerization activity / ATP hydrolysis activity / mitochondrion / ATP binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.27 Å | |||||||||
 Authors Authors | Li Y / Chen YX / Zhou CZ / Hou WT | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: Structural insights into human ABCD3-mediated peroxisomal acyl-CoA translocation. Authors: Yang Li / Zhi-Peng Chen / Da Xu / Liang Wang / Meng-Ting Cheng / Cong-Zhao Zhou / Yuxing Chen / Wen-Tao Hou /  Abstract: Human ABC transporters ABCD1-3 are all localized on the peroxisomal membrane and participate in the β-oxidation of fatty acyl-CoAs, but they differ from each other in substrate specificity. The ...Human ABC transporters ABCD1-3 are all localized on the peroxisomal membrane and participate in the β-oxidation of fatty acyl-CoAs, but they differ from each other in substrate specificity. The transport of branched-chain fatty acids from cytosol to peroxisome is specifically driven by ABCD3, dysfunction of which causes severe liver diseases such as hepatosplenomegaly. Here we report two cryogenic electron microscopy (cryo-EM) structures of ABCD3 bound to phytanoyl-CoA and ATP at resolutions of 2.9 Å and 3.2 Å, respectively. A pair of phytanoyl-CoA molecules were observed in ABCD3, each binding to one transmembrane domain (TMD), which is distinct from our previously reported structure of ABCD1, where each fatty acyl-CoA molecule strongly crosslinks two TMDs. Upon ATP binding, ABCD3 exhibits a conformation that is open towards the peroxisomal matrix, leaving two extra densities corresponding to two CoA molecules deeply embedded in the translocation cavity. Structural analysis combined with substrate-stimulated ATPase activity assays indicated that the present structures might represent two states of ABCD3 in the transport cycle. These findings advance our understanding of fatty acid oxidation and the molecular pathology of related diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39703.map.gz emd_39703.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39703-v30.xml emd-39703-v30.xml emd-39703.xml emd-39703.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39703.png emd_39703.png | 100.4 KB | ||
| Filedesc metadata |  emd-39703.cif.gz emd-39703.cif.gz | 6.4 KB | ||
| Others |  emd_39703_half_map_1.map.gz emd_39703_half_map_1.map.gz emd_39703_half_map_2.map.gz emd_39703_half_map_2.map.gz | 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39703 http://ftp.pdbj.org/pub/emdb/structures/EMD-39703 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39703 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39703 | HTTPS FTP |
-Validation report
| Summary document |  emd_39703_validation.pdf.gz emd_39703_validation.pdf.gz | 805.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39703_full_validation.pdf.gz emd_39703_full_validation.pdf.gz | 805.2 KB | Display | |
| Data in XML |  emd_39703_validation.xml.gz emd_39703_validation.xml.gz | 10.7 KB | Display | |
| Data in CIF |  emd_39703_validation.cif.gz emd_39703_validation.cif.gz | 12.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39703 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39703 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39703 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39703 | HTTPS FTP |
-Related structure data
| Related structure data |  8z0fMC  8z9xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39703.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39703.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
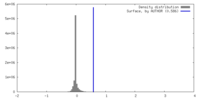
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39703_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
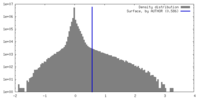
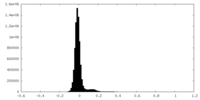
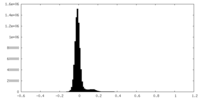
| Density Histograms |
-Half map: #1
| File | emd_39703_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
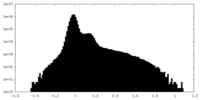
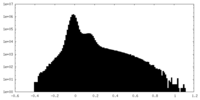
| Density Histograms |
- Sample components
Sample components
-Entire : ATP-bound human peroxisomal ABCD3
| Entire | Name: ATP-bound human peroxisomal ABCD3 |
|---|---|
| Components |
|
-Supramolecule #1: ATP-bound human peroxisomal ABCD3
| Supramolecule | Name: ATP-bound human peroxisomal ABCD3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 140 kDa/nm |
-Macromolecule #1: ATP-binding cassette sub-family D member 3
| Macromolecule | Name: ATP-binding cassette sub-family D member 3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Thioester hydrolases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.678188 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKA VFSKLQLLGQ AIPPKQYAPG VVGMLAVFAL IKLYKQDIRG TKHLVAKTKE GKKERAVVDK VFFSRLIQIL KIMVPRTFC KETGYLVLIA VMLVSRTYCD VWMIQNGTLI ESGIIGRSRK DFKRYLLNFI AAMPLISLVN NFLKYGLNEL K LCFRVRLT ...String: MDYKDDDDKA VFSKLQLLGQ AIPPKQYAPG VVGMLAVFAL IKLYKQDIRG TKHLVAKTKE GKKERAVVDK VFFSRLIQIL KIMVPRTFC KETGYLVLIA VMLVSRTYCD VWMIQNGTLI ESGIIGRSRK DFKRYLLNFI AAMPLISLVN NFLKYGLNEL K LCFRVRLT KYLYEEYLQA FTYYKMGNLD NRIANPDQLL TQDVEKFCNS VVDLYSNLSK PFLDIVLYIF KLTSAIGAQG PA SMMAYLV VSGLFLTRLR RPIGKMTITE QKYEGEYRYV NSRLITNSEE IAFYNGNKRE KQTVHSVFRK LVEHLHNFIL FRF SMGFID SIIAKYLATV VGYLVVSRPF LDLSHPRHLK STHSELLEDY YQSGRMLLRM SQALGRIVLA GREMTRLAGF TARI TELMQ VLKDLNHGKY ERTMVSQQEK GIEGVQVIPL IPGAGEIIIA DNIIKFDHVP LATPNGDVLI RDLNFEVRSG ANVLI CGPN GCGKSSLFRV LGELWPLFGG RLTKPERGKL FYVPQRPYMT LGTLRDQVIY PDGREDQKRK GISDLVLKEY LDNVQL GHI LEREGGWDSV QDWMDVLSGG EKQRMAMARL FYHKPQFAIL DQCTSAVSVD VEGYIYSHCR KVGITLFTVS HRKSLWK HH EYYLHMDGRG NYEFKQITED TVEFGS UniProtKB: ATP-binding cassette sub-family D member 3 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8z0f: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)