[English] 日本語
 Yorodumi
Yorodumi- EMDB-39625: Cryo-EM structure of human mitochondrial pyruvate carrier in comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human mitochondrial pyruvate carrier in complex with the inhibitor UK5099 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitochondrial pyruvate carrier / MPC / pyruvate transport / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate import into mitochondria / inner mitochondrial membrane protein complex / pyruvate transmembrane transporter activity / pyruvate decarboxylation to acetyl-CoA / Pyruvate metabolism / positive regulation of insulin secretion involved in cellular response to glucose stimulus / mitochondrial inner membrane / mitochondrion / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Shi JH / Liang JM / Ma D | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2025 Journal: Nature / Year: 2025Title: Structures and mechanism of the human mitochondrial pyruvate carrier. Authors: Jiaming Liang / Junhui Shi / Ailong Song / Meihua Lu / Kairan Zhang / Meng Xu / Gaoxingyu Huang / Peilong Lu / Xudong Wu / Dan Ma /  Abstract: The mitochondrial pyruvate carrier (MPC) is a mitochondrial inner membrane protein complex that is essential for the uptake of pyruvate into the mitochondrial matrix as the primary carbon source for ...The mitochondrial pyruvate carrier (MPC) is a mitochondrial inner membrane protein complex that is essential for the uptake of pyruvate into the mitochondrial matrix as the primary carbon source for the tricarboxylic acid cycle. Here we present six cryo-electron microscopy structures of human MPC in three states: three structures in the intermembrane space (IMS)-open state, obtained in different conditions; a structure of pyruvate-treated MPC in the occluded state; and two structures in the matrix-facing state, bound with the inhibitor UK5099 or with an inhibitory nanobody on the matrix side. MPC is a heterodimer consisting of MPC1 and MPC2, with the transmembrane domain adopting pseudo-C2 symmetry. Approximate rigid-body movements occur between the IMS-open state and the occluded state, whereas structural changes, mainly on the matrix side, facilitate the transition between the occluded state and the matrix-facing state, revealing an alternating access mechanism during pyruvate transport. In the UK5099-bound structure, the inhibitor fits well and interacts extensively with a pocket that opens to the matrix side. Our findings provide key insights into the mechanisms that underlie MPC-mediated substrate transport, and shed light on the recognition and inhibition of MPC by UK5099, which will facilitate the future development of drugs that target MPC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39625.map.gz emd_39625.map.gz | 117.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39625-v30.xml emd-39625-v30.xml emd-39625.xml emd-39625.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39625.png emd_39625.png | 17.4 KB | ||
| Filedesc metadata |  emd-39625.cif.gz emd-39625.cif.gz | 6.2 KB | ||
| Others |  emd_39625_half_map_1.map.gz emd_39625_half_map_1.map.gz emd_39625_half_map_2.map.gz emd_39625_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39625 http://ftp.pdbj.org/pub/emdb/structures/EMD-39625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39625 | HTTPS FTP |
-Validation report
| Summary document |  emd_39625_validation.pdf.gz emd_39625_validation.pdf.gz | 794.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39625_full_validation.pdf.gz emd_39625_full_validation.pdf.gz | 793.9 KB | Display | |
| Data in XML |  emd_39625_validation.xml.gz emd_39625_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_39625_validation.cif.gz emd_39625_validation.cif.gz | 16.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39625 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39625 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39625 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39625 | HTTPS FTP |
-Related structure data
| Related structure data |  8yw8MC  8yw6C  8yw9C  9knwC  9knxC  9knyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39625.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39625.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.57 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39625_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_39625_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : A membrane transporter complex
| Entire | Name: A membrane transporter complex |
|---|---|
| Components |
|
-Supramolecule #1: A membrane transporter complex
| Supramolecule | Name: A membrane transporter complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Mitochondrial pyruvate carrier 2
| Macromolecule | Name: Mitochondrial pyruvate carrier 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.161725 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSAAGARGLR ATYHRLLDKV ELMLPEKLRP LYNHPAGPRT VFFWAPIMKW GLVCAGLADM ARPAEKLSTA QSAVLMATGF IWSRYSLVI IPKNWSLFAV NFFVGAAGAS QLFRIWRYNQ ELKAKAHKGS DYKDHDGDYK DHDIDYKDDD DK UniProtKB: Mitochondrial pyruvate carrier 2 |
-Macromolecule #2: MPC specific nanobody 1
| Macromolecule | Name: MPC specific nanobody 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.25583 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQAGGSLRL SCAASGFPVT ERVMYWYRQA PGKEREWVAA IDSQGSSTYY ADSVKGRFTI SRDNSKNTVY LQMNSLKPE DTAVYYCKVE VGWGYKGQGT QVTVSSLEHH HHHHHGGSGE QKLISEEDL |
-Macromolecule #3: Mitochondrial pyruvate carrier 1
| Macromolecule | Name: Mitochondrial pyruvate carrier 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.592766 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGALVRKAA DYVRSKDFRD YLMSTHFWGP VANWGLPIAA INDMKKSPEI ISGRMTFALC CYSLTFMRFA YKVQPRNWLL FACHATNEV AQLIQGGRLI KHEMTKTASA GSYPYDVPDY A UniProtKB: Mitochondrial pyruvate carrier 1 |
-Macromolecule #4: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 4 / Number of copies: 1 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Macromolecule #5: (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid
| Macromolecule | Name: (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid / type: ligand / ID: 5 / Number of copies: 1 / Formula: I2R |
|---|---|
| Molecular weight | Theoretical: 288.3 Da |
| Chemical component information | 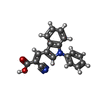 ChemComp-I2R: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































