+Search query
-Structure paper
| Title | Structures and mechanism of the human mitochondrial pyruvate carrier. |
|---|---|
| Journal, issue, pages | Nature, Vol. 641, Issue 8061, Page 258-265, Year 2025 |
| Publish date | Mar 18, 2025 |
 Authors Authors | Jiaming Liang / Junhui Shi / Ailong Song / Meihua Lu / Kairan Zhang / Meng Xu / Gaoxingyu Huang / Peilong Lu / Xudong Wu / Dan Ma /  |
| PubMed Abstract | The mitochondrial pyruvate carrier (MPC) is a mitochondrial inner membrane protein complex that is essential for the uptake of pyruvate into the mitochondrial matrix as the primary carbon source for ...The mitochondrial pyruvate carrier (MPC) is a mitochondrial inner membrane protein complex that is essential for the uptake of pyruvate into the mitochondrial matrix as the primary carbon source for the tricarboxylic acid cycle. Here we present six cryo-electron microscopy structures of human MPC in three states: three structures in the intermembrane space (IMS)-open state, obtained in different conditions; a structure of pyruvate-treated MPC in the occluded state; and two structures in the matrix-facing state, bound with the inhibitor UK5099 or with an inhibitory nanobody on the matrix side. MPC is a heterodimer consisting of MPC1 and MPC2, with the transmembrane domain adopting pseudo-C2 symmetry. Approximate rigid-body movements occur between the IMS-open state and the occluded state, whereas structural changes, mainly on the matrix side, facilitate the transition between the occluded state and the matrix-facing state, revealing an alternating access mechanism during pyruvate transport. In the UK5099-bound structure, the inhibitor fits well and interacts extensively with a pocket that opens to the matrix side. Our findings provide key insights into the mechanisms that underlie MPC-mediated substrate transport, and shed light on the recognition and inhibition of MPC by UK5099, which will facilitate the future development of drugs that target MPC. |
 External links External links |  Nature / Nature /  PubMed:40101766 PubMed:40101766 |
| Methods | EM (single particle) |
| Resolution | 3.01 - 3.72 Å |
| Structure data | EMDB-39624, PDB-8yw6: EMDB-39625, PDB-8yw8: EMDB-39626, PDB-8yw9: EMDB-62464, PDB-9knw: EMDB-62465, PDB-9knx: EMDB-62466, PDB-9kny: |
| Chemicals |  ChemComp-PC8:  ChemComp-CDL: 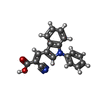 ChemComp-I2R: |
| Source |
|
 Keywords Keywords | PROTEIN TRANSPORT / mitochondrial pyruvate carrier / MPC / pyruvate transport / A membrane transporter complex with substrate |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers















 homo sapiens (human)
homo sapiens (human)
