[English] 日本語
 Yorodumi
Yorodumi- EMDB-39433: structure of phage T6 topoisomerase II ATPase domain bound with AMPPNP -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | structure of phage T6 topoisomerase II ATPase domain bound with AMPPNP | |||||||||
 Map data Map data | T6ATPase half map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | topoisomerase II / ISOMERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationsister chromatid segregation / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity / DNA topoisomerase (ATP-hydrolysing) / DNA topological change / DNA binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  Enterobacteria phage T6 (virus) Enterobacteria phage T6 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | Chen YT / Xin YH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural and functional insights into the T-even type bacteriophage topoisomerase II. Authors: Yuhui Xin / Runqi Xian / Yunge Yang / Jingyuan Cong / Zihe Rao / Xuemei Li / Yutao Chen /  Abstract: T-even type bacteriophages are virulent phages commonly used as model organisms, playing a crucial role in understanding various biological processes. One such process involves the regulation of DNA ...T-even type bacteriophages are virulent phages commonly used as model organisms, playing a crucial role in understanding various biological processes. One such process involves the regulation of DNA topology during phage replication upon host infection, governed by type IIA DNA topoisomerases. In spite of various studies on prokaryotic and eukaryotic counterparts, viral topoisomerase II remains insufficiently understood, especially the unique domain composition of T4 phage. In this study, we determine the cryo-EM structures of topoisomerase II from T4 and T6 phages, including full-length structures of both apo and DNA-binding states which have never been determined before. Together with other conformational states, these structures provide an explicit blueprint of mechanisms of phage topoisomerase II. Particularly, the asymmetric dimeric interactions observed in cryo-EM structures of T6 phage topoisomerase II ATPase domain and central domain bound with DNA shed light on the asynchronous ATP usage and asynchronous cleavage of the G-segment DNA, respectively. The elucidation of phage topoisomerase II's structures and functions not only enhances our understanding of mechanisms and evolutionary parallels with prokaryotic and eukaryotic homologs but also highlights its potential as a model for developing type IIA topoisomerase inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39433.map.gz emd_39433.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39433-v30.xml emd-39433-v30.xml emd-39433.xml emd-39433.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39433.png emd_39433.png | 37.9 KB | ||
| Filedesc metadata |  emd-39433.cif.gz emd-39433.cif.gz | 6 KB | ||
| Others |  emd_39433_half_map_1.map.gz emd_39433_half_map_1.map.gz emd_39433_half_map_2.map.gz emd_39433_half_map_2.map.gz | 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39433 http://ftp.pdbj.org/pub/emdb/structures/EMD-39433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39433 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39433 | HTTPS FTP |
-Validation report
| Summary document |  emd_39433_validation.pdf.gz emd_39433_validation.pdf.gz | 824.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39433_full_validation.pdf.gz emd_39433_full_validation.pdf.gz | 824.5 KB | Display | |
| Data in XML |  emd_39433_validation.xml.gz emd_39433_validation.xml.gz | 10.7 KB | Display | |
| Data in CIF |  emd_39433_validation.cif.gz emd_39433_validation.cif.gz | 12.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39433 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39433 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39433 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39433 | HTTPS FTP |
-Related structure data
| Related structure data |  8yo1MC  8yluC  8yo3C  8yo4C  8yo5C  8yo7C  8yo9C  8yodC  8yonC  9imjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39433.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39433.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T6ATPase half map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
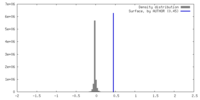
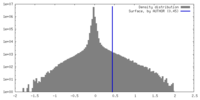
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: T6ATPase half map
| File | emd_39433_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T6ATPase half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
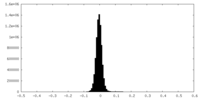
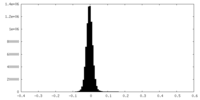
| Density Histograms |
-Half map: T6ATPase half map
| File | emd_39433_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | T6ATPase half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
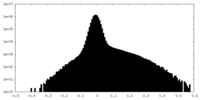
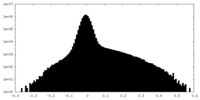
| Density Histograms |
- Sample components
Sample components
-Entire : Phage T6 topoisomerase II ATPase domain bound with AMPPNP
| Entire | Name: Phage T6 topoisomerase II ATPase domain bound with AMPPNP |
|---|---|
| Components |
|
-Supramolecule #1: Phage T6 topoisomerase II ATPase domain bound with AMPPNP
| Supramolecule | Name: Phage T6 topoisomerase II ATPase domain bound with AMPPNP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T6 (virus) Enterobacteria phage T6 (virus) |
-Macromolecule #1: DNA topoisomerase (ATP-hydrolyzing)
| Macromolecule | Name: DNA topoisomerase (ATP-hydrolyzing) / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA topoisomerase (ATP-hydrolysing) |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage T6 (virus) Enterobacteria phage T6 (virus) |
| Molecular weight | Theoretical: 69.237289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIKNEIKILS DIEHIKKRSG MYIGSSANEM HERFLFGKWE SVQYVPGLVK LIDEIIDNSV DEGIRTKFKF ANKINVTIKN NQVTVEDNG RGIPQAMVKT PTGEEIPGPV AAWTIPKAGG NFGDDKERVT GGMNGVGSSL TNIFSVMFVG ETGDGQNNIV V RCSNGMEN ...String: MIKNEIKILS DIEHIKKRSG MYIGSSANEM HERFLFGKWE SVQYVPGLVK LIDEIIDNSV DEGIRTKFKF ANKINVTIKN NQVTVEDNG RGIPQAMVKT PTGEEIPGPV AAWTIPKAGG NFGDDKERVT GGMNGVGSSL TNIFSVMFVG ETGDGQNNIV V RCSNGMEN KSWETIPGKW KGTRVTFIPD FMSFETNELS QVYLDITLDR LQTLAVVYPD IQFTFNGKKV QGNFKKYARQ YD EHAIVQE QENCSIAVGR SPDGFRQLTY VNNIHTKNGG HHIDCVMDDI CEDLIPQIKR KFKIDVTKAR VKECLTIVMF VRD MKNMRF DSQTKERLTS PFGEIRSHIQ LDAKKISRAI LNNEAILMPI IEAALARKLA AEKAAETKAA KKASKAKVHK HIKA NLCGK DADTTLFLTE GDSAIGYLID VRDKELHGGY PLRGKVLNSW GMSYADMLKN KELFDICAIT GLVLGEKAEN LNYHN IAIM TDADHDGLGS IYPSLLGFFS NWPELFEQGR IRFVKTPVII AHVGKKQEWF YTVAEYESAK DALPKHSIRY IKGLGS LEK SEYREMIQNP VYDVVKLPEN WKELFEMLMG DNADLRKEWM SQHHHHHH UniProtKB: DNA topoisomerase (ATP-hydrolyzing) |
-Macromolecule #2: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 2 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)