[English] 日本語
 Yorodumi
Yorodumi- EMDB-39186: Cryo-EM structure of SARS-CoV-2 spike ectodomain (HexaPro, Omicro... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.5 variant) in complex with CeSPIACE, class 2 | ||||||||||||||||||
 Map data Map data | post-processed map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Spike protein / VIRAL PROTEIN-INHIBITOR COMPLEX | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.2 Å | ||||||||||||||||||
 Authors Authors | Suzuki H / Nakamura S / Fujiyoshi Y | ||||||||||||||||||
| Funding support |  Japan, 5 items Japan, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes. Authors: Shun Nakamura / Yukihiro Tanimura / Risa Nomura / Hiroshi Suzuki / Kouki Nishikawa / Akiko Kamegawa / Nobutaka Numoto / Atsushi Tanaka / Shigeru Kawabata / Shoichi Sakaguchi / Akino Emi / ...Authors: Shun Nakamura / Yukihiro Tanimura / Risa Nomura / Hiroshi Suzuki / Kouki Nishikawa / Akiko Kamegawa / Nobutaka Numoto / Atsushi Tanaka / Shigeru Kawabata / Shoichi Sakaguchi / Akino Emi / Youichi Suzuki / Yoshinori Fujiyoshi /  Abstract: Pathogen mutations present an inevitable and challenging problem for therapeutics and the development of mutation-tolerant anti-infective drugs to strengthen global health and combat evolving ...Pathogen mutations present an inevitable and challenging problem for therapeutics and the development of mutation-tolerant anti-infective drugs to strengthen global health and combat evolving pathogens is urgently needed. While spike proteins on viral surfaces are attractive targets for preventing viral entry, they mutate frequently, making it difficult to develop effective therapeutics. Here, we used a structure-guided strategy to engineer an inhibitor peptide against the SARS-CoV-2 spike, called CeSPIACE, with mutation-tolerant and potent binding ability against all variants to enhance affinity for the invariant architecture of the receptor-binding domain (RBD). High-resolution structures of the peptide complexed with mutant RBDs revealed a mechanism of mutation-tolerant inhibition. CeSPIACE bound major mutant RBDs with picomolar affinity and inhibited infection by SARS-CoV-2 variants in VeroE6/TMPRSS2 cells (IC 4 pM to 13 nM) and demonstrated potent in vivo efficacy by inhalation administration in hamsters. Mutagenesis analyses to address mutation risks confirmed tolerance against existing and/or potential future mutations of the RBD. Our strategy of engineering mutation-tolerant inhibitors may be applicable to other infectious diseases. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39186.map.gz emd_39186.map.gz | 140.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39186-v30.xml emd-39186-v30.xml emd-39186.xml emd-39186.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39186_fsc.xml emd_39186_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_39186.png emd_39186.png | 67.4 KB | ||
| Masks |  emd_39186_msk_1.map emd_39186_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-39186.cif.gz emd-39186.cif.gz | 7.9 KB | ||
| Others |  emd_39186_half_map_1.map.gz emd_39186_half_map_1.map.gz emd_39186_half_map_2.map.gz emd_39186_half_map_2.map.gz | 118.3 MB 118.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39186 http://ftp.pdbj.org/pub/emdb/structures/EMD-39186 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39186 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39186 | HTTPS FTP |
-Related structure data
| Related structure data |  8ydzMC  8ydpC  8ydqC  8ydrC  8ydsC  8ydtC  8yduC  8ydvC  8ydwC  8ydxC  8ydyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39186.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39186.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post-processed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.44 Å | ||||||||||||||||||||||||||||||||||||
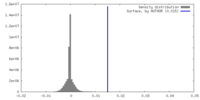
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_39186_msk_1.map emd_39186_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
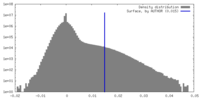
| Density Histograms |
-Half map: half map 1
| File | emd_39186_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_39186_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.5 variant) in co...
| Entire | Name: SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.5 variant) in complex with CeSPIACE |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.5 variant) in co...
| Supramolecule | Name: SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.5 variant) in complex with CeSPIACE type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 Details: the N-terminal sequence (from -18 to 12) is an mammalian secretion signal sequence, and the C-terminal sequence includes "T4 fibritin trimerization mottif" (Gly1211-Leu1237), "HRV 3C ...Details: the N-terminal sequence (from -18 to 12) is an mammalian secretion signal sequence, and the C-terminal sequence includes "T4 fibritin trimerization mottif" (Gly1211-Leu1237), "HRV 3C protease cleavage site" (Leu1241-Pro1248), "8x His tag" (His1250-His1257) and "Twin-strep tag" (Trp1260-Lys1288) Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 144.193672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GSQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF L GVYYHKNN ...String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GSQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF L GVYYHKNN KSWMESEFRV YSSANNCTFE YVSQPFLMDL EGKQGNFKNL REFVFKNIDG YFKIYSKHTP INLVRDLPQG FS ALEPLVD LPIGINITRF QTLLALHRSY LTPGDSSSGW TAGAAAYYVG YLQPRTFLLK YNENGTITDA VDCALDPLSE TKC TLKSFT VEKGIYQTSN FRVQPTESIV RFPNITNLCP FDEVFNATRF ASVYAWNRKR ISNCVADYSV LYNFAPFSAF KCYG VSPTK LNDLCFTNVY ADSFVIRGNE VSQIAPGQTG NIADYNYKLP DDFTGCVIAW NSNKLDSKVG GNYNYRYRLF RKSNL KPFE RDISTEIYQA GNKPCNGVAG VNCYFPLQSY GFRPTYGVGH QPYRVVVLSF ELLHAPATVC GPKKSTNLVK NKCVNF NFN GLTGTGVLTE SNKKFLPFQQ FGRDIADTTD AVRDPQTLEI LDITPCSFGG VSVITPGTNT SNQVAVLYQD VNCTEVP VA IHADQLTPTW RVYSTGSNVF QTRAGCLIGA EHVNNSYECD IPIGAGICAS YQTQTNSPGS ASSVASQSII AYTMSLGA E NSVAYSNNSI AIPTNFTISV TTEILPVSMT KTSVDCTMYI CGDSTECSNL LLQYGSFCTQ LNRALTGIAV EQDKNTQEV FAQVKQIYKT PPIKDFGGFN FSQILPDPSK PSKRSPIEDL LFNKVTLADA GFIKQYGDCL GDIAARDLIC AQKFNGLTVL PPLLTDEMI AQYTSALLAG TITSGWTFGA GPALQIPFPM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS T PSALGKLQ DVVNQNAQAL NTLVKQLSSN FGAISSVLND ILSRLDPPEA EVQIDRLITG RLQSLQTYVT QQLIRAAEIR AS ANLAATK MSECVLGQSK RVDFCGKGYH LMSFPQSAPH GVVFLHVTYV PAQEKNFTTA PAICHDGKAH FPREGVFVSN GTH WFVTQR NFYEPQIITT DNTFVSGNCD VVIGIVNNTV YDPLQPELDS FKEELDKYFK NHTSPDVDLG DISGINASVV NIQK EIDRL NEVAKNLNES LIDLQELGKY EQGSGYIPEA PRDGQAYVRK DGEWVLLSTF LGRSLEVLFQ GPGHHHHHHH HSAWS HPQF EKGGGSGGGG SGGSAWSHPQ FEK UniProtKB: Spike glycoprotein |
-Macromolecule #2: CeSPIACE
| Macromolecule | Name: CeSPIACE / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.721475 KDa |
| Sequence | String: DKLWILQKIY EIMVRLDEEG HGEASLMVSD LIYEFMKRD |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.42 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 69.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 3.4 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: JEOL 3200FSC CRYOHOLDER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)