+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Choline transporter BetT - CHT bound | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | choline transporter / CHOLINE-BINDING PROTEIN | |||||||||
| Function / homology | :  Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas syringae (bacteria) Pseudomonas syringae (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.59 Å | |||||||||
 Authors Authors | Yang TJ / Nian YW / Lin HJ / Li J / Zhang JR / Fan MR | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Structure and mechanism of the osmoregulated choline transporter BetT. Authors: Tianjiao Yang / Yuwei Nian / Huajian Lin / Jing Li / Xiang Lin / Tianming Li / Ruiying Wang / Longfei Wang / Gwyn A Beattie / Jinru Zhang / Minrui Fan /   Abstract: The choline-glycine betaine pathway plays an important role in bacterial survival in hyperosmotic environments. Osmotic activation of the choline transporter BetT promotes the uptake of external ...The choline-glycine betaine pathway plays an important role in bacterial survival in hyperosmotic environments. Osmotic activation of the choline transporter BetT promotes the uptake of external choline for synthesizing the osmoprotective glycine betaine. Here, we report the cryo-electron microscopy structures of BetT in the apo and choline-bound states. Our structure shows that BetT forms a domain-swapped trimer with the C-terminal domain (CTD) of one protomer interacting with the transmembrane domain (TMD) of a neighboring protomer. The substrate choline is bound within a tryptophan prism at the central part of TMD. Together with functional characterization, our results suggest that in species, including the plant pathogen and the human pathogen , BetT is locked at a low-activity state through CTD-mediated autoinhibition in the absence of osmotic stress, and its hyperosmotic activation involves the release of this autoinhibition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39121.map.gz emd_39121.map.gz | 168 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39121-v30.xml emd-39121-v30.xml emd-39121.xml emd-39121.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39121_fsc.xml emd_39121_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_39121.png emd_39121.png | 39.7 KB | ||
| Masks |  emd_39121_msk_1.map emd_39121_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-39121.cif.gz emd-39121.cif.gz | 6.1 KB | ||
| Others |  emd_39121_half_map_1.map.gz emd_39121_half_map_1.map.gz emd_39121_half_map_2.map.gz emd_39121_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39121 http://ftp.pdbj.org/pub/emdb/structures/EMD-39121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39121 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39121 | HTTPS FTP |
-Related structure data
| Related structure data |  8ybqMC  8ybrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39121.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39121.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_39121_msk_1.map emd_39121_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Choline transporter BetT
| Entire | Name: Choline transporter BetT |
|---|---|
| Components |
|
-Supramolecule #1: Choline transporter BetT
| Supramolecule | Name: Choline transporter BetT / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Pseudomonas syringae (bacteria) Pseudomonas syringae (bacteria) |
-Macromolecule #1: BCCT family transporter
| Macromolecule | Name: BCCT family transporter / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas syringae (bacteria) Pseudomonas syringae (bacteria) |
| Molecular weight | Theoretical: 75.25468 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSMSSPSTS SSGTVRMNAP VFYFAASFIL IFGIIVIAFP QASGAWLLAA QNWAANTVGW YYMMVMTLYL VFVVVTALSG FGKIKLGAD HDEPEFSYLS WAGMLFAAGI SITLFFFCVS EPLTHLLQPP QGEGGTAEAA RQGMQLLFLH WGLHGWGVFA F VGMALAYF ...String: MGSMSSPSTS SSGTVRMNAP VFYFAASFIL IFGIIVIAFP QASGAWLLAA QNWAANTVGW YYMMVMTLYL VFVVVTALSG FGKIKLGAD HDEPEFSYLS WAGMLFAAGI SITLFFFCVS EPLTHLLQPP QGEGGTAEAA RQGMQLLFLH WGLHGWGVFA F VGMALAYF AYRHNLPLAL RSALYPLIGK RINGPIGYAV DGFGIIATIF GLGADMGFGV LHLNSGLDYL FGVPHTQWIQ VG LITLMMG AAILVAIAGV DKGVRVMSDI NMLLACALLL FVLFAGPTQH LLNTLVQNIG DYLGALPSKS FDVYAYNKPS DWL GGWTVF YWAWWIAWAP FVGLFIARIS RGRTIREFVF GVLLIPLGFT LAWMSIFGNS AIDQVLNHGM AALGQSAIDD PSMT LYLLL ETYPWSKTVI AVTVFISFVF FVTSADSGTV VLSTLSAKGG NPDEDGPKWL RVFWGVATAL ITSGLLFSGS IDALK SAVV LTSLPFSLIL LLMMWGLHKA FVMESQRQIA QLYSLAPVSG SRRGGWRQRL SQAVHYPSRD EVYRFLDQTV RPAIDE VTA VFVEKGLNVV NVPDPSNDSV TLEIGHGEER PFIYQVQMKG FFTPSFARGG MGSKQLNNRR YYRAEVHLSE GSQDYDL VG YTKEQVINDV LDQYERHMQF LHLVRGSGSG SGSGSWSHPQ FEK UniProtKB: UNIPROTKB: A0A2S3V5U4 |
-Macromolecule #2: CHOLINE ION
| Macromolecule | Name: CHOLINE ION / type: ligand / ID: 2 / Number of copies: 3 / Formula: CHT |
|---|---|
| Molecular weight | Theoretical: 104.171 Da |
| Chemical component information | 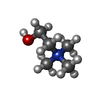 ChemComp-CHT: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 66 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





























