+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Ebola virus nucleocapsid subunit | |||||||||||||||||||||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN / VIRAL PROTEIN-RNA complex | |||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endomembrane system / viral RNA genome packaging / helical viral capsid / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / viral budding from plasma membrane / viral nucleocapsid / host cell cytoplasm / symbiont-mediated suppression of host innate immune response / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / ribonucleoprotein complex ...host cell endomembrane system / viral RNA genome packaging / helical viral capsid / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / viral budding from plasma membrane / viral nucleocapsid / host cell cytoplasm / symbiont-mediated suppression of host innate immune response / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / ribonucleoprotein complex / host cell plasma membrane / virion membrane / structural molecule activity / RNA binding / membrane Similarity search - Function | |||||||||||||||||||||||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Fujita-Fujiharu Y / Hu S / Hirabayashi A / Takamatsu Y / Ng Y / Houri K / Muramoto Y / Nakano M / Sugita Y / Noda T | |||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  Japan, 14 items Japan, 14 items
| |||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Structural basis for Ebola virus nucleocapsid assembly and function regulated by VP24. Authors: Yoko Fujita-Fujiharu / Shangfan Hu / Ai Hirabayashi / Yuki Takamatsu / Yen Ni Ng / Kazuya Houri / Yukiko Muramoto / Masahiro Nakano / Yukihiko Sugita / Takeshi Noda /   Abstract: The Ebola virus, a member of the Filoviridae family, causes severe hemorrhagic fever in humans. Filamentous virions contain a helical nucleocapsid responsible for genome transcription, replication, ...The Ebola virus, a member of the Filoviridae family, causes severe hemorrhagic fever in humans. Filamentous virions contain a helical nucleocapsid responsible for genome transcription, replication, and packaging into progeny virions. The nucleocapsid consists of a helical nucleoprotein (NP)-viral genomic RNA complex forming the core structure, to which VP24 and VP35 bind externally. Two NPs, each paired with a VP24 molecule, constitute a repeating unit. However, the detailed nucleocapsid structure remains unclear. Here, we determine the nucleocapsid-like structure within virus-like particles at 4.6 Å resolution using single-particle cryo-electron microscopy. Mutational analysis identifies specific interactions between the two NPs and two VP24s and demonstrates that each of the two VP24s in different orientations distinctively regulates nucleocapsid assembly, viral RNA synthesis, intracellular transport of the nucleocapsid, and infectious virion production. Our findings highlight the sophisticated mechanisms underlying the assembly and functional regulation of the nucleocapsid and provide insights into antiviral development. | |||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39081.map.gz emd_39081.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39081-v30.xml emd-39081-v30.xml emd-39081.xml emd-39081.xml | 30.1 KB 30.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39081_fsc.xml emd_39081_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_39081.png emd_39081.png | 67.7 KB | ||
| Masks |  emd_39081_msk_1.map emd_39081_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-39081.cif.gz emd-39081.cif.gz | 8.2 KB | ||
| Others |  emd_39081_additional_1.map.gz emd_39081_additional_1.map.gz emd_39081_half_map_1.map.gz emd_39081_half_map_1.map.gz emd_39081_half_map_2.map.gz emd_39081_half_map_2.map.gz | 404.4 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39081 http://ftp.pdbj.org/pub/emdb/structures/EMD-39081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39081 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39081 | HTTPS FTP |
-Validation report
| Summary document |  emd_39081_validation.pdf.gz emd_39081_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39081_full_validation.pdf.gz emd_39081_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_39081_validation.xml.gz emd_39081_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_39081_validation.cif.gz emd_39081_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39081 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39081 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39081 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39081 | HTTPS FTP |
-Related structure data
| Related structure data |  8y9jMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39081.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39081.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2375 Å | ||||||||||||||||||||||||||||||||||||
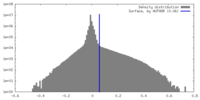
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_39081_msk_1.map emd_39081_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
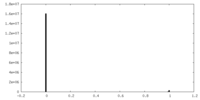
| Density Histograms |
-Additional map: #1
| File | emd_39081_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
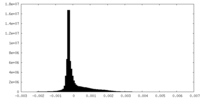
| Density Histograms |
-Half map: #2
| File | emd_39081_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
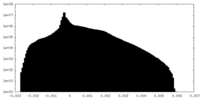
| Density Histograms |
-Half map: #1
| File | emd_39081_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Zaire ebolavirus
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Zaire ebolavirus
| Supramolecule | Name: Zaire ebolavirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 186538 / Sci species name: Zaire ebolavirus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 83.3875 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDSRPQKIWM APSLTESDMD YHKILTAGLS VQQGIVRQRV IPVYQVNNLE EICQLIIQAF EAGVDFQESA DSFLLMLCLH HAYQGDYKL FLESGAVKYL EGHGFRFEVK KRDGVKRLEE LLPAVSSGKN IKRTLAAMPE EETTEANAGQ FLSFASLFLP K LVVGEKAC ...String: MDSRPQKIWM APSLTESDMD YHKILTAGLS VQQGIVRQRV IPVYQVNNLE EICQLIIQAF EAGVDFQESA DSFLLMLCLH HAYQGDYKL FLESGAVKYL EGHGFRFEVK KRDGVKRLEE LLPAVSSGKN IKRTLAAMPE EETTEANAGQ FLSFASLFLP K LVVGEKAC LEKVQRQIQV HAEQGLIQYP TAWQSVGHMM VIFRLMRTNF LIKFLLIHQG MHMVAGHDAN DAVISNSVAQ AR FSGLLIV KTVLDHILQK TERGVRLHPL ARTAKVKNEV NSFKAALSSL AKHGEYAPFA RLLNLSGVNN LEHGLFPQLS AIA LGVATA HGSTLAGVNV GEQYQQLREA ATEAEKQLQQ YAESRELDHL GLDDQEKKIL MNFHQKKNEI SFQQTNAMVT LRKE RLAKL TEAITAASLP KTSGHYDDDD DIPFPGPIND DDNPGHQDDD PTDSQDTTIP DVVVDPDDGS YGEYQSYSEN GMNAP DDLV LFDLDEDDED TKPVPNRSTK GGQQKNSQKG QHIEGRQTQS RPIQNVPGPH RTIHHASAPL TDNDRRNEPS GSTSPR MLT PINEEADPLD DADDETSSLP PLESDDEEQD RDGTSNRTPT VAPPAPVYRD HSEKKELPQD EQQDQDHTQE ARNQDSD NT QSEHSFEEMY RHILRSQGPF DAVLYYHMMK DEPVVFSTSD GKEYTYPDSL EEEYPPWLTE KEAMNEENRF VTLDGQQF Y WPVMNHKNKF MAILQHHQ UniProtKB: Nucleoprotein |
-Macromolecule #2: Membrane-associated protein VP24
| Macromolecule | Name: Membrane-associated protein VP24 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.250811 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAKATGRYNL ISPKKDLEKG VVLSDLCNFL VSQTIQGWKV YWAGIEFDVT HKGMALLHRL KTNDFAPAWS MTRNLFPHLF QNPNSTIES PLWALRVILA AGIQDQLIDQ SLIEPLAGAL GLISDWLLTT NTNHFNMRTQ RVKEQLSLKM LSLIRSNILK F INKLDALH ...String: MAKATGRYNL ISPKKDLEKG VVLSDLCNFL VSQTIQGWKV YWAGIEFDVT HKGMALLHRL KTNDFAPAWS MTRNLFPHLF QNPNSTIES PLWALRVILA AGIQDQLIDQ SLIEPLAGAL GLISDWLLTT NTNHFNMRTQ RVKEQLSLKM LSLIRSNILK F INKLDALH VVNYNGLLSS IEIGTQNHTI IITRTNMGFL VELQEPDKSA MNRMKPGPAK FSLLHESTLK AFTQGSSTRM QS LILEFNS SLAI UniProtKB: Membrane-associated protein VP24 |
-Macromolecule #3: RNA (12-MER)
| Macromolecule | Name: RNA (12-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Kidney Homo sapiens (human) / Organ: Kidney |
| Molecular weight | Theoretical: 3.629032 KDa |
| Sequence | String: UUUUUUUUUU UU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Pressure: 0.02 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281.15 K / Instrument: FEI VITROBOT MARK IV Details: The sample was applied to both sides of the grid. The grids were blotted for 14 seconds.. | ||||||||||||
| Details | The total 2.5 microlitre sample was applied to both sides of the glow-discharged grid. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Microscope was modified with a Cs corrector (CEOS GmbH) Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 5897 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Protocol: FLEXIBLE FIT | ||||||||
| Output model |  PDB-8y9j: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)