[English] 日本語
 Yorodumi
Yorodumi- EMDB-39079: Versatile Aromatic Prenyltransferase auraA in complex with DMSPP ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Versatile Aromatic Prenyltransferase auraA in complex with DMSPP and cyclo-(L-Val-DH-His) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Prenyltransferase / Imidazole-Containing Diketopiperazines / TRANSFERASE | ||||||||||||
| Biological species |  Penicillium (fungus) Penicillium (fungus) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.67 Å | ||||||||||||
 Authors Authors | Li D / Zhang Y / Wang W / Wang P | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Characterization and structural analysis of a versatile aromatic prenyltransferase for imidazole-containing diketopiperazines. Authors: Wenxue Wang / Peng Wang / Chuanteng Ma / Kang Li / Zian Wang / Yuting Liu / Lu Wang / Guojian Zhang / Qian Che / Tianjiao Zhu / Yuzhong Zhang / Dehai Li /  Abstract: Prenylation modifications of natural products play essential roles in chemical diversity and bioactivities, but imidazole modification prenyltransferases are not well investigated. Here, we discover ...Prenylation modifications of natural products play essential roles in chemical diversity and bioactivities, but imidazole modification prenyltransferases are not well investigated. Here, we discover a dimethylallyl tryptophan synthase family prenyltransferase, AuraA, that catalyzes the rare dimethylallylation on the imidazole moiety in the biosynthesis of aurantiamine. Biochemical assays validate that AuraA could accept both cyclo-(L-Val-L-His) and cyclo-(L-Val-DH-His) as substrates, while the prenylation modes are completely different, yielding C2-regular and C5-reverse products, respectively. Cryo-electron microscopy analysis of AuraA and its two ternary complex structures reveal two distinct modes for receptor binding, demonstrating a tolerance for altered orientations of highly similar receptors. The mutation experiments further demonstrate the promiscuity of AuraA towards imidazole-C-dimethylallylation. In this work, we also characterize a case of AuraA mutant-catalyzed dimethylallylation of imidazole moiety, offering available structural insights into the utilization and engineering of dimethylallyl tryptophan synthase family prenyltransferases. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39079.map.gz emd_39079.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39079-v30.xml emd-39079-v30.xml emd-39079.xml emd-39079.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39079_fsc.xml emd_39079_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_39079.png emd_39079.png | 63 KB | ||
| Filedesc metadata |  emd-39079.cif.gz emd-39079.cif.gz | 5.9 KB | ||
| Others |  emd_39079_half_map_1.map.gz emd_39079_half_map_1.map.gz emd_39079_half_map_2.map.gz emd_39079_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39079 http://ftp.pdbj.org/pub/emdb/structures/EMD-39079 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39079 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39079 | HTTPS FTP |
-Related structure data
| Related structure data |  8y9gMC  8y9dC  8y9eC  9jhxC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39079.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39079.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_39079_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_39079_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Versatile Aromatic Prenyltransferase auraA in complex with DMSPP ...
| Entire | Name: Versatile Aromatic Prenyltransferase auraA in complex with DMSPP and cyclo-(L-Val-DH-His) |
|---|---|
| Components |
|
-Supramolecule #1: Versatile Aromatic Prenyltransferase auraA in complex with DMSPP ...
| Supramolecule | Name: Versatile Aromatic Prenyltransferase auraA in complex with DMSPP and cyclo-(L-Val-DH-His) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Penicillium (fungus) Penicillium (fungus) |
-Macromolecule #1: Versatile Aromatic Prenyltransferase auraA
| Macromolecule | Name: Versatile Aromatic Prenyltransferase auraA / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Penicillium (fungus) Penicillium (fungus) |
| Molecular weight | Theoretical: 46.433684 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SPSPWDFLGR VLQFQHGDHK RWWDVLAPVF GISMASIGYK LDVQYRHLLV LYDAVIPNMG PFPNSNASNI TWTSPFPPGP LEASVNYQA GESSMFRFTI EPVGPHAGTP ADPVNELAAK QLMQRLGQLQ PGGVDSTMFD HFYPLLCVDG PEARRQWDSI A HIYHKCHT ...String: SPSPWDFLGR VLQFQHGDHK RWWDVLAPVF GISMASIGYK LDVQYRHLLV LYDAVIPNMG PFPNSNASNI TWTSPFPPGP LEASVNYQA GESSMFRFTI EPVGPHAGTP ADPVNELAAK QLMQRLGQLQ PGGVDSTMFD HFYPLLCVDG PEARRQWDSI A HIYHKCHT VTALDMQRSA ACTLKTYFPP LLRSTIMNTS MVDIMFDAVE SFRKQSGLYF DYTKIKEFMS EEKTHETMMV DR SYLSFDC LDPAKSRIKI YTEAKVKTLE EAYSFWSLGG RLSGPEIDYG FKIVSQMWDA IYSKELPGGK QRENNHIQIN WEM SAKDSS VAPKLYLTVI EDYDAYVSSA IVDLFTGLGW AAHVQTHKKI EKEAYPMCDA NPQSTHAYVW ISLAYKKTGP YITV YTNPG ASILE |
-Macromolecule #2: DIMETHYLALLYL S-THIOLODIPHOSPHATE
| Macromolecule | Name: DIMETHYLALLYL S-THIOLODIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 4 / Formula: DST |
|---|---|
| Molecular weight | Theoretical: 262.158 Da |
| Chemical component information | 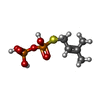 ChemComp-DST: |
-Macromolecule #3: (3~{Z},6~{S})-3-(1~{H}-imidazol-4-ylmethylidene)-6-propan-2-yl-pi...
| Macromolecule | Name: (3~{Z},6~{S})-3-(1~{H}-imidazol-4-ylmethylidene)-6-propan-2-yl-piperazine-2,5-dione type: ligand / ID: 3 / Number of copies: 4 / Formula: A1LYF |
|---|---|
| Molecular weight | Theoretical: 234.254 Da |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 49.62 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































