[English] 日本語
 Yorodumi
Yorodumi- EMDB-39029: Structure of the ige-fc bound to its high affinity receptor fc(ep... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the ige-fc bound to its high affinity receptor fc(epsilon)ri | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / antibody / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationFc epsilon receptor (FCERI) signaling / FCERI mediated NF-kB activation / Dectin-2 family / Role of LAT2/NTAL/LAB on calcium mobilization / Platelet Adhesion to exposed collagen / serotonin secretion / GPVI-mediated activation cascade / IgE receptor activity / Fc-epsilon receptor I complex / Cell surface interactions at the vascular wall ...Fc epsilon receptor (FCERI) signaling / FCERI mediated NF-kB activation / Dectin-2 family / Role of LAT2/NTAL/LAB on calcium mobilization / Platelet Adhesion to exposed collagen / serotonin secretion / GPVI-mediated activation cascade / IgE receptor activity / Fc-epsilon receptor I complex / Cell surface interactions at the vascular wall / FCERI mediated Ca+2 mobilization / Fc receptor mediated stimulatory signaling pathway / T cell differentiation involved in immune response / high-affinity IgE receptor activity / negative regulation of mast cell apoptotic process / IgE B cell receptor complex / type I hypersensitivity / mast cell activation / mast cell apoptotic process / Fc-gamma receptor III complex / FCERI mediated MAPK activation / positive regulation of interleukin-3 production / eosinophil degranulation / serotonin secretion by platelet / neutrophil activation involved in immune response / positive regulation of mast cell degranulation / positive regulation of mast cell cytokine production / positive regulation of type III hypersensitivity / Fc-gamma receptor signaling pathway / regulation of platelet activation / positive regulation of type IIa hypersensitivity / IgE binding / leukotriene biosynthetic process / positive regulation of type I hypersensitivity / regulation of release of sequestered calcium ion into cytosol / interleukin-3-mediated signaling pathway / positive regulation of protein localization to cell surface / positive regulation of granulocyte macrophage colony-stimulating factor production / type 2 immune response / IgG binding / immunoglobulin receptor binding / Neutrophil degranulation / phagocytosis, engulfment / mast cell degranulation / antigen processing and presentation of exogenous peptide antigen via MHC class I / Fc-epsilon receptor signaling pathway / positive regulation of interleukin-4 production / immunoglobulin mediated immune response / positive regulation of interleukin-10 production / cellular response to low-density lipoprotein particle stimulus / regulation of immune response / neutrophil chemotaxis / positive regulation of calcium-mediated signaling / SH2 domain binding / osteoclast differentiation / B cell differentiation / positive regulation of phagocytosis / integrin-mediated signaling pathway / B cell receptor signaling pathway / protein localization to plasma membrane / peptide antigen assembly with MHC class II protein complex / MHC class II protein complex / phosphoprotein binding / establishment of localization in cell / calcium-mediated signaling / antigen processing and presentation of exogenous peptide antigen via MHC class II / positive regulation of immune response / peptide antigen binding / positive regulation of T cell activation / receptor internalization / positive regulation of interleukin-6 production / positive regulation of tumor necrosis factor production / MHC class II protein complex binding / late endosome membrane / cell surface receptor signaling pathway / endosome / defense response to bacterium / immune response / protein heterodimerization activity / innate immune response / external side of plasma membrane / lysosomal membrane / protein kinase binding / cell surface / signal transduction / protein homodimerization activity / extracellular region / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Du S / Deng MJ / Xiao JY | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structural insights into the high-affinity IgE receptor FcεRI complex. Authors: Meijie Deng / Shuo Du / Handi Hou / Junyu Xiao /  Abstract: Immunoglobulin E (IgE) plays a pivotal role in allergic responses. The high-affinity IgE receptor, FcεRI, found on mast cells and basophils, is central to the effector functions of IgE. FcεRI is a ...Immunoglobulin E (IgE) plays a pivotal role in allergic responses. The high-affinity IgE receptor, FcεRI, found on mast cells and basophils, is central to the effector functions of IgE. FcεRI is a tetrameric complex, comprising FcεRIα, FcεRIβ and a homodimer of FcRγ (originally known as FcεRIγ), with FcεRIα recognizing the Fc region of IgE (Fcε) and FcεRIβ-FcRγ facilitating signal transduction. Additionally, FcRγ is a crucial component of other immunoglobulin receptors, including those for IgG (FcγRI and FcγRIIIA) and IgA (FcαRI). However, the molecular basis of FcεRI assembly and the structure of FcRγ have remained elusive. Here we elucidate the cryogenic electron microscopy structure of the Fcε-FcεRI complex. FcεRIα has an essential role in the receptor's assembly, interacting with FcεRIβ and both FcRγ subunits. FcεRIβ is structured as a compact four-helix bundle, similar to the B cell antigen CD20. The FcRγ dimer exhibits an asymmetric architecture, and coils with the transmembrane region of FcεRIα to form a three-helix bundle. A cholesterol-like molecule enhances the interaction between FcεRIβ and the FcεRIα-FcRγ complex. Our mutagenesis analyses further indicate similarities between the interaction of FcRγ with FcεRIα and FcγRIIIA, but differences in that with FcαRI. These findings deepen our understanding of the signalling mechanisms of FcεRI and offer insights into the functionality of other immune receptors dependent on FcRγ. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39029.map.gz emd_39029.map.gz | 79 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39029-v30.xml emd-39029-v30.xml emd-39029.xml emd-39029.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39029.png emd_39029.png | 24.4 KB | ||
| Filedesc metadata |  emd-39029.cif.gz emd-39029.cif.gz | 6.8 KB | ||
| Others |  emd_39029_half_map_1.map.gz emd_39029_half_map_1.map.gz emd_39029_half_map_2.map.gz emd_39029_half_map_2.map.gz | 77.6 MB 77.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39029 http://ftp.pdbj.org/pub/emdb/structures/EMD-39029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39029 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39029 | HTTPS FTP |
-Related structure data
| Related structure data |  8y81MC  8y84C  8z0tC  8zgsC  8zgtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39029.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39029.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
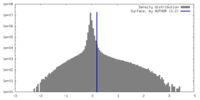
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_39029_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_39029_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of the ige-fc bound to its high affinity receptor fc(ep...
| Entire | Name: Structure of the ige-fc bound to its high affinity receptor fc(epsilon)ri |
|---|---|
| Components |
|
-Supramolecule #1: Structure of the ige-fc bound to its high affinity receptor fc(ep...
| Supramolecule | Name: Structure of the ige-fc bound to its high affinity receptor fc(epsilon)ri type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: High affinity immunoglobulin epsilon receptor subunit alpha
| Macromolecule | Name: High affinity immunoglobulin epsilon receptor subunit alpha type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.83016 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDTGGSARLC LALVLISLGV MLTATQKSVV SLDPPWIRIL TGDKVTLICN GNNSSQMNST KWIHNDSISN VKSSHWVIVS ATIQDSGKY ICQKQGFYKS KPVYLNVMQE WLLLQSSADV VLDNGSFDIR CRSWKKWKVH KVIYYKDDIA FKYSYDSNNI S IRKATFND ...String: MDTGGSARLC LALVLISLGV MLTATQKSVV SLDPPWIRIL TGDKVTLICN GNNSSQMNST KWIHNDSISN VKSSHWVIVS ATIQDSGKY ICQKQGFYKS KPVYLNVMQE WLLLQSSADV VLDNGSFDIR CRSWKKWKVH KVIYYKDDIA FKYSYDSNNI S IRKATFND SGSYHCTGYL NKVECKSDKF SIAVVKDYTI EYRWLQLIFP SLAVILFAVD TGLWFSTHKQ FESILKIQKT GK GKKKG UniProtKB: High affinity immunoglobulin epsilon receptor subunit alpha |
-Macromolecule #2: High affinity immunoglobulin epsilon receptor subunit beta
| Macromolecule | Name: High affinity immunoglobulin epsilon receptor subunit beta type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.747752 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDTENKSRAD LALPNPQESP SAPDIELLEA SPPAKALPEK PASPPPQQTW QSFLKKELEF LGVTQVLVGL ICLCFGTVVC STLQTSDFD DEVLLLYRAG YPFWGAVLFV LSGFLSIMSE RKNTLYLVRG SLGANIVSSI AAGLGIAILI LNLSNNSAYM N YCKDITED ...String: MDTENKSRAD LALPNPQESP SAPDIELLEA SPPAKALPEK PASPPPQQTW QSFLKKELEF LGVTQVLVGL ICLCFGTVVC STLQTSDFD DEVLLLYRAG YPFWGAVLFV LSGFLSIMSE RKNTLYLVRG SLGANIVSSI AAGLGIAILI LNLSNNSAYM N YCKDITED DGCFVTSFIT ELVLMLLFLT ILAFCSAVLL IIYRIGQEFE RSKVPDDRLY EELHVYSPIY SALEDTREAS AP VVS UniProtKB: High affinity immunoglobulin epsilon receptor subunit beta |
-Macromolecule #3: High affinity immunoglobulin epsilon receptor subunit gamma
| Macromolecule | Name: High affinity immunoglobulin epsilon receptor subunit gamma type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.459315 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIPAVILFLL LLVEEAAALG EPQLCYILDA ILFLYGIVLT LLYCRLKIQV RKADIASREK SDAVYTGLNT RNQETYETLK HEKPPQGSG WSHPQFEKGS GDYKDDDDKG SGWSHPQFEK UniProtKB: High affinity immunoglobulin epsilon receptor subunit gamma |
-Macromolecule #4: Immunoglobulin heavy constant epsilon
| Macromolecule | Name: Immunoglobulin heavy constant epsilon / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.815438 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSVPTQVLGL LLLWLTDARC DIARPVNITK PTVDLLHSSC DPNAFHSTIQ LYCFVYGHIQ NDVSIHWLMD DRKIYETHAQ NVLIKEEGK LASTYSRLNI TQQQWMSEST FTCKVTSQGE NYWAHTRRCS DDEPRGVITY LIPPSPLDLY ENGTPKLTCL V LDLESEEN ...String: MSVPTQVLGL LLLWLTDARC DIARPVNITK PTVDLLHSSC DPNAFHSTIQ LYCFVYGHIQ NDVSIHWLMD DRKIYETHAQ NVLIKEEGK LASTYSRLNI TQQQWMSEST FTCKVTSQGE NYWAHTRRCS DDEPRGVITY LIPPSPLDLY ENGTPKLTCL V LDLESEEN ITVTWVRERK KSIGSASQRS TKHHNATTSI TSILPVDAKD WIEGEGYQCR VDHPHFPKPI VRSITKAPGK RS APEVYVF LPPEEEEKDK RTLTCLIQNF FPEDISVQWL QDSKLIPKSQ HSTTTPLKYN GSNQRFFIFS RLEVTKALWT QTK QFTCRV IHEALREPRK LERTISKSLG NTSLRPSQAS MHHHHHH UniProtKB: Immunoglobulin heavy constant epsilon |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #8: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 8 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)