+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the auto-inhibited Dark monomer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dark / apoptosis / cryo-EM | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of humoral immune response / positive regulation of glial cell apoptotic process / Formation of apoptosome / salivary gland histolysis / melanization defense response / sarcosine catabolic process / Activation of caspases through apoptosome-mediated cleavage / Regulation of the apoptosome activity / compound eye retinal cell programmed cell death / central nervous system formation ...negative regulation of humoral immune response / positive regulation of glial cell apoptotic process / Formation of apoptosome / salivary gland histolysis / melanization defense response / sarcosine catabolic process / Activation of caspases through apoptosome-mediated cleavage / Regulation of the apoptosome activity / compound eye retinal cell programmed cell death / central nervous system formation / positive regulation of apoptotic process involved in development / chaeta development / sperm individualization / apoptosome / autophagic cell death / Neutrophil degranulation / S-adenosylmethionine cycle / CARD domain binding / programmed cell death / triglyceride homeostasis / response to starvation / dendrite morphogenesis / response to gamma radiation / neuron cellular homeostasis / ADP binding / apoptotic process / structural molecule activity / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
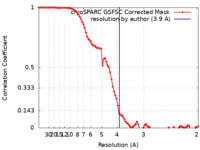
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Tian L / Li Y / Shi Y | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Dark and Dronc activation in . Authors: Lu Tian / Yini Li / Yigong Shi /  Abstract: The onset of apoptosis is characterized by a cascade of caspase activation, where initiator caspases are activated by a multimeric adaptor complex known as the apoptosome. In , the initiator caspase ...The onset of apoptosis is characterized by a cascade of caspase activation, where initiator caspases are activated by a multimeric adaptor complex known as the apoptosome. In , the initiator caspase Dronc undergoes autocatalytic activation in the presence of the Dark apoptosome. Despite rigorous investigations, the activation mechanism for Dronc remains elusive. Here, we report the cryo-EM structures of an auto-inhibited Dark monomer and a single-layered, multimeric Dark/Dronc complex. Our biochemical analysis suggests that the auto-inhibited Dark oligomerizes upon binding to Dronc, which is sufficient for the activation of both Dark and Dronc. In contrast, the previously observed double-ring Dark apoptosome may represent a non-functional or "off-pathway" conformation. These findings expand our understanding on the molecular mechanism of apoptosis in . | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38994.map.gz emd_38994.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38994-v30.xml emd-38994-v30.xml emd-38994.xml emd-38994.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38994_fsc.xml emd_38994_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_38994.png emd_38994.png | 116.7 KB | ||
| Filedesc metadata |  emd-38994.cif.gz emd-38994.cif.gz | 6 KB | ||
| Others |  emd_38994_half_map_1.map.gz emd_38994_half_map_1.map.gz emd_38994_half_map_2.map.gz emd_38994_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38994 http://ftp.pdbj.org/pub/emdb/structures/EMD-38994 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38994 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38994 | HTTPS FTP |
-Validation report
| Summary document |  emd_38994_validation.pdf.gz emd_38994_validation.pdf.gz | 1016.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38994_full_validation.pdf.gz emd_38994_full_validation.pdf.gz | 1016 KB | Display | |
| Data in XML |  emd_38994_validation.xml.gz emd_38994_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_38994_validation.cif.gz emd_38994_validation.cif.gz | 20.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38994 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38994 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38994 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38994 | HTTPS FTP |
-Related structure data
| Related structure data |  8y6pMC  8y6qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38994.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38994.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38994_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_38994_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of the auto-inhibited Dark monomer
| Entire | Name: Structure of the auto-inhibited Dark monomer |
|---|---|
| Components |
|
-Supramolecule #1: Structure of the auto-inhibited Dark monomer
| Supramolecule | Name: Structure of the auto-inhibited Dark monomer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Apaf-1 related killer DARK
| Macromolecule | Name: Apaf-1 related killer DARK / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 166.603641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDFETGEHQY QYKDILSVFE DAFVDNFDCK DVQDMPKSIL SKEEIDHIIM SKDAVSGTLR LFWTLLSKQE EMVQKFVEEV LRINYKFLM SPIKTEQRQP SMMTRMYIEQ RDRLYNDNQV FAKYNVSRLQ PYLKLRQALL ELRPAKNVLI DGVLGSGKTW V ALDVCLSY ...String: MDFETGEHQY QYKDILSVFE DAFVDNFDCK DVQDMPKSIL SKEEIDHIIM SKDAVSGTLR LFWTLLSKQE EMVQKFVEEV LRINYKFLM SPIKTEQRQP SMMTRMYIEQ RDRLYNDNQV FAKYNVSRLQ PYLKLRQALL ELRPAKNVLI DGVLGSGKTW V ALDVCLSY KVQCKMDFKI FWLNLKNCNS PETVLEMLQK LLYQIDPNWT SRSDHSSNIK LRIHSIQAEL RRLLKSKPYE NC LLVLLNV QNAKAWNAFN LSCKILLTTR FKQVTDFLSA ATTTHISLDH HSMTLTPDEV KSLLLKYLDC RPQDLPREVL TTN PRRLSI IAESIRAGLA TWDNWKHVNC DKLTTIIESS LNVLEPAEYR KMFDRLSVFP PSAHIPTILL SLIWFDVIKS DVMV VVNKL HKYSLVEKQP KESTISIPSI YLELKVKLEN EYALHRSIVD HYNIPKTFDS DDLIPPYLDQ YFYSHIGHHL KNIEH PERM TLFRMVFLDF RFLEQKIRHD STAWNASGSI LNTLQQLKFY KPYICDNDPK YERLVNAILD FLPKIEENLI CSKYTD LLR IALMAEDEAI FEEAHKQVQR FDDRVWFTNH GRFHQHRQII NLGDNEGRHA VYLHNDFCLI ALASGQILLT DVSLEGE DT YLLRDESDSS DILRMAVFNQ QKHLITLHCN GSVKLWSLWP DCPGRRHSGG SKQQLVNSVV KRFIGSYANL KIVAFYLN E DAGLPEANIQ LHVAFINGDV SILNWDEQDQ EFKLSHVPVL KTMQSGIRCF VQVLKRYYVV CTSNCTLTVW DLTNGSSNT LELHVFNVEN DTPLALDVFD ERSKTATVLL IFKYSVWRLN FLPGLSVSLQ SEAVQLPEGS FITCGKRSTD GRYLLLGTSE GLIVYDLKI SDPVLRSNVS EHIECVDIYE LFDPVYKYIV LCGAKGKQVV HVHTLRSVSG SNSHQNREIA WVHSADEISV M TKACLEPN VYLRSLMDMT RERTQLLAVD SKERIHLIKP AISRISEWST ITPTHAASNC KINAISAFND EQIFVGYVDG VI IDVIHDT ALPQQFIEEP IDYLKQVSPN ILVASAHSAQ KTVIFQLEKI DPLQPNDQWP LMMDVSTKYA SLQEGQYIIL FSD HGVCHL DIANPSAFVK PKDSEEYIVG FDLKNSLLFL AYENNIIDVF RLIFSCNQLR YEQICEEEIA QKAKISYLVA TDDG TMLAM GFENGTLELF AVENRKVQLI YSIEEVHEHC IRQLLFSPCK LLLISCAEQL CFWNVTHMRN NQLEREQKRR RSRRH KQHS VTQEDAVDAA PIAADIDVDV TFVADEFHPV NRGTAELWRN KRGNAIRPEL LACVKFVGNE ARQFFTDAHF SHFYAI DDE GVYYHLQLLE LSRLQPPPDP VTLDIANQYE DLKNLRILDS PLMQDSDSEG ADVVGNLVLE KNGGVARATP ILEEASS HH HHHH UniProtKB: Apoptosome protein Dark |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)