+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Post-translocated Pol IV transcription elongation complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRANSCRIPTION/DNA/RNA / elongation / Pol IV / RNA polymerase / TRANSCRIPTION-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationstomatal complex patterning / siRNA-mediated long-distance post-transcriptional gene silencing / RNA polymerase IV complex / transposable element silencing by siRNA-mediated heterochromatin formation / RNA polymerase V complex / gene silencing by siRNA-directed DNA methylation / stomatal complex development / siRNA transcription / DNA/RNA hybrid binding / regulatory ncRNA-mediated post-transcriptional gene silencing ...stomatal complex patterning / siRNA-mediated long-distance post-transcriptional gene silencing / RNA polymerase IV complex / transposable element silencing by siRNA-mediated heterochromatin formation / RNA polymerase V complex / gene silencing by siRNA-directed DNA methylation / stomatal complex development / siRNA transcription / DNA/RNA hybrid binding / regulatory ncRNA-mediated post-transcriptional gene silencing / siRNA processing / regulatory ncRNA-mediated gene silencing / plastid / RNA polymerase complex / regulation of immune response / defense response to fungus / heterochromatin / RNA polymerase II, core complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / nucleic acid binding / transcription by RNA polymerase II / protein dimerization activity / single-stranded RNA binding / RNA-directed RNA polymerase / nucleotide binding / RNA-directed RNA polymerase activity / DNA-templated transcription / nucleolus / mitochondrion / DNA binding / zinc ion binding / nucleoplasm / metal ion binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
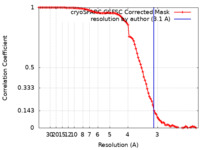
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Huang K / Fang CL / Zhang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Transcription elongation of the plant RNA polymerase IV is prone to backtracking. Authors: Chengli Fang / Kun Huang / Xiaoxian Wu / Hongwei Zhang / Zhanxi Gu / Jiawei Wang / Yu Zhang /  Abstract: RNA polymerase IV (Pol IV) forms a complex with RNA-directed RNA polymerase 2 (RDR2) to produce double-stranded RNA (dsRNA) precursors essential for plant gene silencing. In the "backtracking- ...RNA polymerase IV (Pol IV) forms a complex with RNA-directed RNA polymerase 2 (RDR2) to produce double-stranded RNA (dsRNA) precursors essential for plant gene silencing. In the "backtracking-triggered RNA channeling" model, Pol IV backtracks and delivers its transcript's 3' terminus to RDR2, which synthesizes dsRNA. However, the mechanisms underlying Pol IV backtracking and RNA protection from cleavage are unclear. Here, we determined cryo-electron microscopy structures of Pol IV elongation complexes at four states of its nucleotide addition cycle (NAC): posttranslocation, guanosine triphosphate-bound, pretranslocation, and backtracked states. The structures reveal that Pol IV maintains an open DNA cleft and kinked bridge helix in all NAC states, loosely interacts with the nucleoside triphosphate substrate, and barely contacts proximal backtracked nucleotides. Biochemical data indicate that Pol IV is inefficient in forward translocation and RNA cleavage. These findings suggest that Pol IV transcription elongation is prone to backtracking and incapable of RNA hydrolysis, ensuring efficient dsRNA production by Pol IV-RDR2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38471.map.gz emd_38471.map.gz | 45.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38471-v30.xml emd-38471-v30.xml emd-38471.xml emd-38471.xml | 32 KB 32 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38471_fsc.xml emd_38471_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_38471.png emd_38471.png | 76.8 KB | ||
| Filedesc metadata |  emd-38471.cif.gz emd-38471.cif.gz | 10.1 KB | ||
| Others |  emd_38471_half_map_1.map.gz emd_38471_half_map_1.map.gz emd_38471_half_map_2.map.gz emd_38471_half_map_2.map.gz | 84.6 MB 84.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38471 http://ftp.pdbj.org/pub/emdb/structures/EMD-38471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38471 | HTTPS FTP |
-Validation report
| Summary document |  emd_38471_validation.pdf.gz emd_38471_validation.pdf.gz | 891.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38471_full_validation.pdf.gz emd_38471_full_validation.pdf.gz | 891.4 KB | Display | |
| Data in XML |  emd_38471_validation.xml.gz emd_38471_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_38471_validation.cif.gz emd_38471_validation.cif.gz | 23.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38471 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38471 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38471 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38471 | HTTPS FTP |
-Related structure data
| Related structure data |  8xmcMC  8xmbC  8xmdC  8xmeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38471.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38471.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
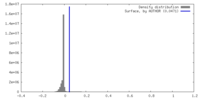
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38471_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
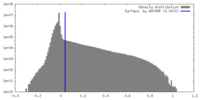
| Density Histograms |
-Half map: #1
| File | emd_38471_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
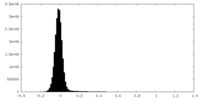
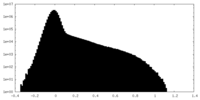
| Density Histograms |
- Sample components
Sample components
+Entire : Post-translocated Pol IV transcription elongation complex
+Supramolecule #1: Post-translocated Pol IV transcription elongation complex
+Macromolecule #1: DNA-directed RNA polymerase IV subunit 1
+Macromolecule #2: DNA-directed RNA polymerases IV and V subunit 2
+Macromolecule #3: DNA-directed RNA polymerases II, IV and V subunit 3
+Macromolecule #4: DNA-directed RNA polymerases IV and V subunit 4
+Macromolecule #5: DNA-directed RNA polymerases II and IV subunit 5A
+Macromolecule #6: DNA-directed RNA polymerases II, IV and V subunit 6A
+Macromolecule #7: DNA-directed RNA polymerase IV subunit 7
+Macromolecule #8: DNA-directed RNA polymerases II, IV and V subunit 8B
+Macromolecule #9: DNA-directed RNA polymerases II, IV and V subunit 9A
+Macromolecule #10: DNA-directed RNA polymerases II, IV and V subunit 10
+Macromolecule #11: DNA-directed RNA polymerases II, IV and V subunit 11
+Macromolecule #12: DNA-directed RNA polymerases II, IV and V subunit 12
+Macromolecule #13: RNA-dependent RNA polymerase 2
+Macromolecule #14: Non-template DNA
+Macromolecule #16: Template DNA
+Macromolecule #15: RNA
+Macromolecule #17: ZINC ION
+Macromolecule #18: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)