+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human pyruvate carboxylase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Biotin-dependent carboxylase / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpyruvate carboxylase / pyruvate carboxylase activity / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism / NADP+ metabolic process / viral RNA genome packaging / pyruvate metabolic process / host-mediated activation of viral process / Pyruvate metabolism / : ...pyruvate carboxylase / pyruvate carboxylase activity / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism / NADP+ metabolic process / viral RNA genome packaging / pyruvate metabolic process / host-mediated activation of viral process / Pyruvate metabolism / : / Gluconeogenesis / biotin binding / viral release from host cell / gluconeogenesis / lipid metabolic process / mitochondrial matrix / negative regulation of gene expression / mitochondrion / ATP binding / metal ion binding / identical protein binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.61 Å | |||||||||
 Authors Authors | Zhou FY / Zhang YY / Zhou Q / Hu Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC). Authors: Zhou FY / Zhang YY / Zhu Y / Zhou Q / Shi Y / Hu Q | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38442.map.gz emd_38442.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38442-v30.xml emd-38442-v30.xml emd-38442.xml emd-38442.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38442.png emd_38442.png | 34.3 KB | ||
| Filedesc metadata |  emd-38442.cif.gz emd-38442.cif.gz | 5.5 KB | ||
| Others |  emd_38442_half_map_1.map.gz emd_38442_half_map_1.map.gz emd_38442_half_map_2.map.gz emd_38442_half_map_2.map.gz | 59 MB 59 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38442 http://ftp.pdbj.org/pub/emdb/structures/EMD-38442 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38442 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38442 | HTTPS FTP |
-Related structure data
| Related structure data |  8xl9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38442.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38442.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0773 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38442_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_38442_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homotetramer complex of human pyruvate carboxylase
| Entire | Name: Homotetramer complex of human pyruvate carboxylase |
|---|---|
| Components |
|
-Supramolecule #1: Homotetramer complex of human pyruvate carboxylase
| Supramolecule | Name: Homotetramer complex of human pyruvate carboxylase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Pyruvate carboxylase, mitochondrial
| Macromolecule | Name: Pyruvate carboxylase, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: pyruvate carboxylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 129.799359 KDa |
| Sequence | String: MLKFRTVHGG LRLLGIRRTS TAPAASPNVR RLEYKPIKKV MVANRGEIAI RVFRACTELG IRTVAIYSEQ DTGQMHRQKA DEAYLIGRG LAPVQAYLHI PDIIKVAKEN NVDAVHPGYG FLSERADFAQ ACQDAGVRFI GPSPEVVRKM GDKVEARAIA I AAGVPVVP ...String: MLKFRTVHGG LRLLGIRRTS TAPAASPNVR RLEYKPIKKV MVANRGEIAI RVFRACTELG IRTVAIYSEQ DTGQMHRQKA DEAYLIGRG LAPVQAYLHI PDIIKVAKEN NVDAVHPGYG FLSERADFAQ ACQDAGVRFI GPSPEVVRKM GDKVEARAIA I AAGVPVVP GTDAPITSLH EAHEFSNTYG FPIIFKAAYG GGGRGMRVVH SYEELEENYT RAYSEALAAF GNGALFVEKF IE KPRHIEV QILGDQYGNI LHLYERDCSI QRRHQKVVEI APAAHLDPQL RTRLTSDSVK LAKQVGYENA GTVEFLVDRH GKH YFIEVN SRLQVEHTVT EEITDVDLVH AQIHVAEGRS LPDLGLRQEN IRINGCAIQC RVTTEDPARS FQPDTGRIEV FRSG EGMGI RLDNASAFQG AVISPHYDSL LVKVIAHGKD HPTAATKMSR ALAEFRVRGV KTNIAFLQNV LNNQQFLAGT VDTQF IDEN PELFQLRPAQ NRAQKLLHYL GHVMVNGPTT PIPVKASPSP TDPVVPAVPI GPPPAGFRDI LLREGPEGFA RAVRNH PGL LLMDTTFRDA HQSLLATRVR THDLKKIAPY VAHNFSKLFS MENWGGATFD VAMRFLYECP WRRLQELREL IPNIPFQ ML LRGANAVGYT NYPDNVVFKF CEVAKENGMD VFRVFDSLNY LPNMLLGMEA AGSAGGVVEA AISYTGDVAD PSRTKYSL Q YYMGLAEELV RAGTHILCIK DMAGLLKPTA CTMLVSSLRD RFPDLPLHIH THDTSGAGVA AMLACAQAGA DVVDVAADS MSGMTSQPSM GALVACTRGT PLDTEVPMER VFDYSEYWEG ARGLYAAFDC TATMKSGNSD VYENEIPGGQ YTNLHFQAHS MGLGSKFKE VKKAYVEANQ MLGDLIKVTP SSKIVGDLAQ FMVQNGLSRA EAEAQAEELS FPRSVVEFLQ GYIGVPHGGF P EPFRSKVL KDLPRVEGRP GASLPPLDLQ ALEKELVDRH GEEVTPEDVL SAAMYPDVFA HFKDFTATFG PLDSLNTRLF LQ GPKIAEE FEVELERGKT LHIKALAVSD LNRAGQRQVF FELNGQLRSI LVKDTQAMKE MHFHPKALKD VKGQIGAPMP GKV IDIKVV AGAKVAKGQP LCVLSAMKME TVVTSPMEGT VRKVHVTKDM TLEGDDLILE IE UniProtKB: Pyruvate carboxylase, mitochondrial |
-Macromolecule #2: BIOTIN
| Macromolecule | Name: BIOTIN / type: ligand / ID: 2 / Number of copies: 4 / Formula: BTN |
|---|---|
| Molecular weight | Theoretical: 244.311 Da |
| Chemical component information | 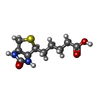 ChemComp-BTN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.61 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 677493 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































