[English] 日本語
 Yorodumi
Yorodumi- EMDB-3830: Shape-programmable self-limiting hierarchical self-assembly of fi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3830 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Shape-programmable self-limiting hierarchical self-assembly of finite-size objects through accurate DNA design: V-brick 2.0 for the assembly into 2D finite-size rings | |||||||||
 Map data Map data | V-brick 2.0 for the assembly into 2D finite-size rings and higher-order tube-like objects. | |||||||||
 Sample Sample |
| |||||||||
| Biological species | synthetic construct (others) | |||||||||
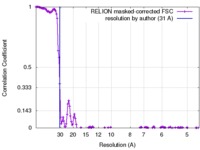
| Method | single particle reconstruction / cryo EM / Resolution: 31.0 Å | |||||||||
 Authors Authors | Dietz H / Wagenbauer KF | |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Gigadalton-scale shape-programmable DNA assemblies. Authors: Klaus F Wagenbauer / Christian Sigl / Hendrik Dietz /  Abstract: Natural biomolecular assemblies such as molecular motors, enzymes, viruses and subcellular structures often form by self-limiting hierarchical oligomerization of multiple subunits. Large structures ...Natural biomolecular assemblies such as molecular motors, enzymes, viruses and subcellular structures often form by self-limiting hierarchical oligomerization of multiple subunits. Large structures can also assemble efficiently from a few components by combining hierarchical assembly and symmetry, a strategy exemplified by viral capsids. De novo protein design and RNA and DNA nanotechnology aim to mimic these capabilities, but the bottom-up construction of artificial structures with the dimensions and complexity of viruses and other subcellular components remains challenging. Here we show that natural assembly principles can be combined with the methods of DNA origami to produce gigadalton-scale structures with controlled sizes. DNA sequence information is used to encode the shapes of individual DNA origami building blocks, and the geometry and details of the interactions between these building blocks then control their copy numbers, positions and orientations within higher-order assemblies. We illustrate this strategy by creating planar rings of up to 350 nanometres in diameter and with atomic masses of up to 330 megadaltons, micrometre-long, thick tubes commensurate in size to some bacilli, and three-dimensional polyhedral assemblies with sizes of up to 1.2 gigadaltons and 450 nanometres in diameter. We achieve efficient assembly, with yields of up to 90 per cent, by using building blocks with validated structure and sufficient rigidity, and an accurate design with interaction motifs that ensure that hierarchical assembly is self-limiting and able to proceed in equilibrium to allow for error correction. We expect that our method, which enables the self-assembly of structures with sizes approaching that of viruses and cellular organelles, can readily be used to create a range of other complex structures with well defined sizes, by exploiting the modularity and high degree of addressability of the DNA origami building blocks used. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3830.map.gz emd_3830.map.gz | 23.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3830-v30.xml emd-3830-v30.xml emd-3830.xml emd-3830.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3830_fsc.xml emd_3830_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3830.png emd_3830.png | 40.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3830 http://ftp.pdbj.org/pub/emdb/structures/EMD-3830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3830 | HTTPS FTP |
-Related structure data
| Related structure data |  3826C  3827C  3828C  3829C  3831C  3832C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3830.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3830.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | V-brick 2.0 for the assembly into 2D finite-size rings and higher-order tube-like objects. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.319 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
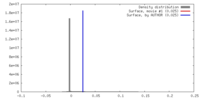
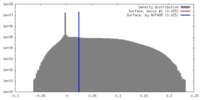
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA origami
| Entire | Name: DNA origami |
|---|---|
| Components |
|
-Supramolecule #1: DNA origami
| Supramolecule | Name: DNA origami / type: complex / ID: 1 / Parent: 0 / Details: Includes two M13-derived genomic scaffold chains |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 11 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)