[English] 日本語
 Yorodumi
Yorodumi- EMDB-3812: Tomogram of e. coli carrying the ple7 plasmid carrying YFP-MreB h... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3812 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tomogram of e. coli carrying the ple7 plasmid carrying YFP-MreB hyper-overexpressed by induction with 1uM IPTG | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | electron tomography | |||||||||
 Authors Authors | Swulius MT / Jensen GJ | |||||||||
 Citation Citation |  Journal: J Bacteriol / Year: 2012 Journal: J Bacteriol / Year: 2012Title: The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-Terminal yellow fluorescent protein tag. Authors: Matthew T Swulius / Grant J Jensen /  Abstract: Based on fluorescence microscopy, the actin homolog MreB has been thought to form extended helices surrounding the cytoplasm of rod-shaped bacterial cells. The presence of these and other putative ...Based on fluorescence microscopy, the actin homolog MreB has been thought to form extended helices surrounding the cytoplasm of rod-shaped bacterial cells. The presence of these and other putative helices has come to dominate models of bacterial cell shape regulation, chromosome segregation, polarity, and motility. Here we use electron cryotomography to show that MreB does in fact form extended helices and filaments in Escherichia coli when yellow fluorescent protein (YFP) is fused to its N terminus but native (untagged) MreB expressed to the same levels does not. In contrast, mCherry fused to an internal loop (MreB-RFP(SW)) does not induce helices. The helices are therefore an artifact of the placement of the fluorescent protein tag. YFP-MreB helices were also clearly distinguishable from the punctate, "patchy" localization patterns of MreB-RFP(SW), even by standard light microscopy. The many interpretations in the literature of such punctate patterns as helices should therefore be reconsidered. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3812.map.gz emd_3812.map.gz | 417.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3812-v30.xml emd-3812-v30.xml emd-3812.xml emd-3812.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3812.png emd_3812.png | 119.6 KB | ||
| Others |  emd_3812_additional_1.map.gz emd_3812_additional_1.map.gz emd_3812_additional_2.map.gz emd_3812_additional_2.map.gz emd_3812_additional_3.map.gz emd_3812_additional_3.map.gz emd_3812_additional_4.map.gz emd_3812_additional_4.map.gz | 555.3 MB 517.2 MB 462.4 MB 630.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3812 http://ftp.pdbj.org/pub/emdb/structures/EMD-3812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3812 | HTTPS FTP |
-Validation report
| Summary document |  emd_3812_validation.pdf.gz emd_3812_validation.pdf.gz | 153.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3812_full_validation.pdf.gz emd_3812_full_validation.pdf.gz | 152.5 KB | Display | |
| Data in XML |  emd_3812_validation.xml.gz emd_3812_validation.xml.gz | 4.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3812 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3812 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3812 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3812 | HTTPS FTP |
-Related structure data
| Related structure data |  3810C  3811C C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-10115 (Title: Tilt-series of e. coli carrying the ple7 plasmid carrying YFP-MreB hyper-overexpressed by induction with 1uM IPTG EMPIAR-10115 (Title: Tilt-series of e. coli carrying the ple7 plasmid carrying YFP-MreB hyper-overexpressed by induction with 1uM IPTGData size: 5.0 Data #1: Tilt-series for e. coli carrying the ple7 plasmid carrying YFP-MreB hyper-overexpressed by induction with 1uM IPTG [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3812.map.gz / Format: CCP4 / Size: 984.4 MB / Type: IMAGE STORED AS SIGNED BYTE Download / File: emd_3812.map.gz / Format: CCP4 / Size: 984.4 MB / Type: IMAGE STORED AS SIGNED BYTE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 9.46 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: #1
| File | emd_3812_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
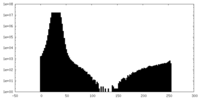
| Density Histograms |
-Additional map: #2
| File | emd_3812_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #3
| File | emd_3812_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #4
| File | emd_3812_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia Coli
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia Coli
| Supramolecule | Name: Escherichia Coli / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Helical MreB Cytoskeleton
| Supramolecule | Name: Helical MreB Cytoskeleton / type: organelle_or_cellular_component / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
 Processing Processing | electron tomography |
|---|---|
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Concentration: 50.0 ug/ml / Component - Formula: C16H19N3O4S / Component - Name: Ampicillin Details: MC1000, MC1000/pLE6, and MC1000/pLE7 were grown in LB at 37C with 50ug/ml ampicillin when appropriate. |
|---|---|
| Grid | Model: Quantifoil R2/2 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Max: 123.15 K |
| Specialist optics | Energy filter - Name: FEI |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 1016 pixel / Digitization - Dimensions - Height: 1016 pixel / Average electron dose: 0.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Software - Name:  IMOD IMODSoftware - details: Tomograms were reconstructed and modelled Number images used: 501 |
|---|
 Movie
Movie Controller
Controller


 Z
Z Y
Y X
X