+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human XPR1 in open state | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transport open state / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphate transmembrane transporter activity / phosphate ion transport / intracellular phosphate ion homeostasis / phosphate ion transmembrane transport / cellular response to phosphate starvation / inositol hexakisphosphate binding / efflux transmembrane transporter activity / response to virus / virus receptor activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.61 Å | |||||||||
 Authors Authors | Jiang DH / Yan R | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Human XPR1 structures reveal phosphate export mechanism. Authors: Rui Yan / Huiwen Chen / Chuanyu Liu / Jun Zhao / Di Wu / Juquan Jiang / Jianke Gong / Daohua Jiang /  Abstract: Inorganic phosphate (Pi) is a fundamental macronutrient for all living organisms, the homeostasis of which is critical for numerous biological activities. As the only known human Pi exporter to date, ...Inorganic phosphate (Pi) is a fundamental macronutrient for all living organisms, the homeostasis of which is critical for numerous biological activities. As the only known human Pi exporter to date, XPR1 has an indispensable role in cellular Pi homeostasis. Dysfunction of XPR1 is associated with neurodegenerative disease. However, the mechanisms underpinning XPR1-mediated Pi efflux and regulation by the intracellular inositol polyphosphate (InsPP) sensor SPX domain remain poorly understood. Here we present cryo-electron microscopy structures of human XPR1 in Pi-bound closed, open and InsP-bound forms, revealing the structural basis for XPR1 gating and regulation by InsPPs. XPR1 consists of an N-terminal SPX domain, a dimer-formation core domain and a Pi transport domain. Within the transport domain, three basic clusters are responsible for Pi binding and transport, and a conserved W573 acts as a molecular switch for gating. In addition, the SPX domain binds to InsP and facilitates Pi efflux by liberating the C-terminal loop that limits Pi entry. This study provides a conceptual framework for the mechanistic understanding of Pi homeostasis by XPR1 homologues in fungi, plants and animals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38067.map.gz emd_38067.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38067-v30.xml emd-38067-v30.xml emd-38067.xml emd-38067.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38067.png emd_38067.png | 32.8 KB | ||
| Filedesc metadata |  emd-38067.cif.gz emd-38067.cif.gz | 6.3 KB | ||
| Others |  emd_38067_half_map_1.map.gz emd_38067_half_map_1.map.gz emd_38067_half_map_2.map.gz emd_38067_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38067 http://ftp.pdbj.org/pub/emdb/structures/EMD-38067 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38067 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38067 | HTTPS FTP |
-Validation report
| Summary document |  emd_38067_validation.pdf.gz emd_38067_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38067_full_validation.pdf.gz emd_38067_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_38067_validation.xml.gz emd_38067_validation.xml.gz | 13.8 KB | Display | |
| Data in CIF |  emd_38067_validation.cif.gz emd_38067_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38067 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38067 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38067 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38067 | HTTPS FTP |
-Related structure data
| Related structure data |  8x5eMC  8x5bC  8x5fC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38067.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38067.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_38067_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
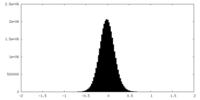
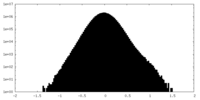
| Density Histograms |
-Half map: half map
| File | emd_38067_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
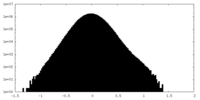
| Density Histograms |
- Sample components
Sample components
-Entire : transport
| Entire | Name: transport |
|---|---|
| Components |
|
-Supramolecule #1: transport
| Supramolecule | Name: transport / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 53 member 1
| Macromolecule | Name: Solute carrier family 53 member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.831422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: APAWTTFRVG LFCGIFIVLN ITLVLAAVFK LETDRSIWPL IRIYRGGFLL IEFLFLLGIN TYGWRQAGVN HVLIFELNPR SNLSHQHLF EIAGFLGILW CLSLLACFFA PISVIPTYVY PLALYGFMVF FLINPTKTFY YKSRFWLLKL LFRVFTAPFH K VGFADFWL ...String: APAWTTFRVG LFCGIFIVLN ITLVLAAVFK LETDRSIWPL IRIYRGGFLL IEFLFLLGIN TYGWRQAGVN HVLIFELNPR SNLSHQHLF EIAGFLGILW CLSLLACFFA PISVIPTYVY PLALYGFMVF FLINPTKTFY YKSRFWLLKL LFRVFTAPFH K VGFADFWL ADQLNSLSVI LMDLEYMICF YSLELKWDES KGLLPNNSEE SGICHKYTYG VRAIVQCIPA WLRFIQCLRR YR DTKRAFP HLVNAGKYST TFFMVTFAAL YSTHKERGHS DTMVFFYLWI VFYIISSCYT LIWDLKMDWG LFDKNAGENT FLR EEIVYP QKAYYYCAII EDVILRFAWT IQISITSTTL LPHSGDIIAT VFAPLEVFRR FVWNFFRLEN EHLNNCGEFR AVRD ISVAP LNADDQTLLE QMMDQDDGVR NRQKNRSWKY NQSISLRRPR LASQSKARDT KVLIEDTDDE ANT UniProtKB: Solute carrier family 53 member 1 |
-Macromolecule #2: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #3: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyl...
| Macromolecule | Name: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate type: ligand / ID: 3 / Number of copies: 6 / Formula: 6OU |
|---|---|
| Molecular weight | Theoretical: 717.996 Da |
| Chemical component information |  ChemComp-6OU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)