[English] 日本語
 Yorodumi
Yorodumi- EMDB-37848: RNA polymerase II elongation complex bound with Elf1, Spt4/5 and ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(0) of the nucleosome | |||||||||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | Transcription / RNAPII / Nucleosome / Cryo-EM | |||||||||||||||||||||||||||||||||
| Biological species |  Komagataella phaffii (fungus) / Komagataella phaffii (fungus) /  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.7 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Akatsu M / Fujita R / Ogasawara M / Ehara H / Kujirai T / Takizawa Y / Sekine S / Kurumizaka H | |||||||||||||||||||||||||||||||||
| Funding support |  Japan, 10 items Japan, 10 items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2023 Journal: J Biol Chem / Year: 2023Title: Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA. Authors: Munetaka Akatsu / Haruhiko Ehara / Tomoya Kujirai / Risa Fujita / Tomoko Ito / Ken Osumi / Mitsuo Ogasawara / Yoshimasa Takizawa / Shun-Ichi Sekine / Hitoshi Kurumizaka /  Abstract: RNA polymerase II (RNAPII) transcribes DNA wrapped in the nucleosome by stepwise pausing, especially at nucleosomal superhelical locations -5 and -1 [SHL(-5) and SHL(-1), respectively]. In the ...RNA polymerase II (RNAPII) transcribes DNA wrapped in the nucleosome by stepwise pausing, especially at nucleosomal superhelical locations -5 and -1 [SHL(-5) and SHL(-1), respectively]. In the present study, we performed cryo-electron microscopy analyses of RNAPII-nucleosome complexes paused at a major nucleosomal pausing site, SHL(-1). We determined two previously undetected structures, in which the transcribed DNA behind RNAPII is sharply kinked at the RNAPII exit tunnel and rewrapped around the nucleosomal histones in front of RNAPII by DNA looping. This DNA kink shifts the DNA orientation toward the nucleosome, and the transcribed DNA region interacts with basic amino acid residues of histones H2A, H2B, and H3 exposed by the RNAPII-mediated nucleosomal DNA peeling. The DNA loop structure was not observed in the presence of the transcription elongation factors Spt4/5 and Elf1. These RNAPII-nucleosome structures provide important information for understanding the functional relevance of DNA looping during transcription elongation in the nucleosome. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37848.map.gz emd_37848.map.gz | 12.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37848-v30.xml emd-37848-v30.xml emd-37848.xml emd-37848.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37848.png emd_37848.png | 92 KB | ||
| Filedesc metadata |  emd-37848.cif.gz emd-37848.cif.gz | 4.5 KB | ||
| Others |  emd_37848_half_map_1.map.gz emd_37848_half_map_1.map.gz emd_37848_half_map_2.map.gz emd_37848_half_map_2.map.gz | 140.7 MB 141.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37848 http://ftp.pdbj.org/pub/emdb/structures/EMD-37848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37848 | HTTPS FTP |
-Validation report
| Summary document |  emd_37848_validation.pdf.gz emd_37848_validation.pdf.gz | 873.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37848_full_validation.pdf.gz emd_37848_full_validation.pdf.gz | 873.4 KB | Display | |
| Data in XML |  emd_37848_validation.xml.gz emd_37848_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  emd_37848_validation.cif.gz emd_37848_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37848 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37848 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37848 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37848 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37848.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37848.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_37848_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
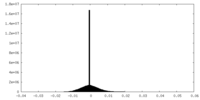
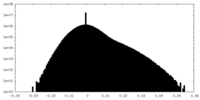
| Density Histograms |
-Half map: #2
| File | emd_37848_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
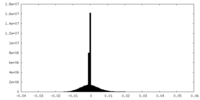
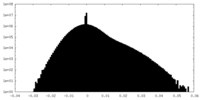
| Density Histograms |
- Sample components
Sample components
-Entire : RNA polymerase II - nucleosome complex
| Entire | Name: RNA polymerase II - nucleosome complex |
|---|---|
| Components |
|
-Supramolecule #1: RNA polymerase II - nucleosome complex
| Supramolecule | Name: RNA polymerase II - nucleosome complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#22 |
|---|
-Supramolecule #2: RNA polymerase II
| Supramolecule | Name: RNA polymerase II / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii (fungus) Komagataella phaffii (fungus) |
-Supramolecule #3: Histones
| Supramolecule | Name: Histones / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: DNA, RNA
| Supramolecule | Name: DNA, RNA / type: complex / ID: 4 / Parent: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.09144 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 20 mM HEPES-KOH(pH7.5), 50 mM Potassium acetate, 200 nM Zinc acetate, 0.1 mM TCEP-HCl | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III | |||||||||||||||
| Details | dsDNA concentration is 0.09144 mg/mL. This sample contains 0.005% Tween20. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 59.84 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 12176 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)