[English] 日本語
 Yorodumi
Yorodumi- EMDB-37752: Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | E3 ligase / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of free ubiquitin chain polymerization / response to mitochondrial depolarisation / positive regulation of mitophagy / positive regulation by virus of viral protein levels in host cell / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / neural tube development / positive regulation of regulatory T cell differentiation / biological process involved in interaction with symbiont ...positive regulation of free ubiquitin chain polymerization / response to mitochondrial depolarisation / positive regulation of mitophagy / positive regulation by virus of viral protein levels in host cell / spindle assembly involved in female meiosis / epigenetic programming in the zygotic pronuclei / UV-damage excision repair / neural tube development / positive regulation of regulatory T cell differentiation / biological process involved in interaction with symbiont / negative regulation of cardiac muscle cell apoptotic process / regulation of mitotic cell cycle phase transition / WD40-repeat domain binding / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / Macroautophagy / negative regulation of reproductive process / negative regulation of developmental process / regulation of G1/S transition of mitotic cell cycle / cullin family protein binding / viral release from host cell / protein phosphatase activator activity / axoneme / autophagosome assembly / ectopic germ cell programmed cell death / ubiquitin-like ligase-substrate adaptor activity / positive regulation of viral genome replication / proteasomal protein catabolic process / mitophagy / phagocytic vesicle / positive regulation of autophagy / positive regulation of gluconeogenesis / autophagosome / cellular response to starvation / Recognition of DNA damage by PCNA-containing replication complex / DNA Damage Recognition in GG-NER / nucleotide-excision repair / Dual Incision in GG-NER / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / Formation of TC-NER Pre-Incision Complex / Formation of Incision Complex in GG-NER / regulation of circadian rhythm / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / Wnt signaling pathway / protein polyubiquitination / positive regulation of protein catabolic process / cellular response to UV / rhythmic process / site of double-strand break / Neddylation / GTPase binding / ubiquitin-dependent protein catabolic process / protein phosphatase binding / protein-macromolecule adaptor activity / damaged DNA binding / mitochondrial outer membrane / proteasome-mediated ubiquitin-dependent protein catabolic process / negative regulation of neuron apoptotic process / cell differentiation / cytoskeleton / chromosome, telomeric region / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / protein ubiquitination / negative regulation of cell population proliferation / focal adhesion / DNA repair / intracellular membrane-bounded organelle / ubiquitin protein ligase binding / apoptotic process / DNA damage response / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / protein-containing complex binding / nucleolus / perinuclear region of cytoplasm / endoplasmic reticulum / protein-containing complex / mitochondrion / extracellular space / DNA binding / extracellular exosome / nucleoplasm / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.08 Å | |||||||||
 Authors Authors | Liu M / Wang Y / Su MY / Stjepanovic G | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation. Authors: Ming Liu / Yang Wang / Fei Teng / Xinyi Mai / Xi Wang / Ming-Yuan Su / Goran Stjepanovic /  Abstract: AMBRA1 is a tumor suppressor protein that functions as a substrate receptor of the ubiquitin conjugation system with roles in autophagy and the cell cycle regulatory network. The intrinsic disorder ...AMBRA1 is a tumor suppressor protein that functions as a substrate receptor of the ubiquitin conjugation system with roles in autophagy and the cell cycle regulatory network. The intrinsic disorder of AMBRA1 has thus far precluded its structural determination. To solve this problem, we analyzed the dynamics of AMBRA1 using hydrogen deuterium exchange mass spectrometry (HDX-MS). The HDX results indicated that AMBRA1 is a highly flexible protein and can be stabilized upon interaction with DDB1, the adaptor of the Cullin4A/B E3 ligase. Here, we present the cryo-EM structure of AMBRA1 in complex with DDB1 at 3.08 Å resolution. The structure shows that parts of the N- and C-terminal structural regions in AMBRA1 fold together into the highly dynamic WD40 domain and reveals how DDB1 engages with AMBRA1 to create a binding scaffold for substrate recruitment. The N-terminal helix-loop-helix motif and WD40 domain of AMBRA1 associate with the double-propeller fold of DDB1. We also demonstrate that DDB1 binding-defective AMBRA1 mutants prevent ubiquitination of the substrate Cyclin D1 in vitro and increase cell cycle progression. Together, these results provide structural insights into the AMBRA1-ubiquitin ligase complex and suggest a mechanism by which AMBRA1 acts as a hub involved in various physiological processes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37752.map.gz emd_37752.map.gz | 50.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37752-v30.xml emd-37752-v30.xml emd-37752.xml emd-37752.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37752.png emd_37752.png | 38 KB | ||
| Filedesc metadata |  emd-37752.cif.gz emd-37752.cif.gz | 6.7 KB | ||
| Others |  emd_37752_half_map_1.map.gz emd_37752_half_map_1.map.gz emd_37752_half_map_2.map.gz emd_37752_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37752 http://ftp.pdbj.org/pub/emdb/structures/EMD-37752 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37752 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37752 | HTTPS FTP |
-Validation report
| Summary document |  emd_37752_validation.pdf.gz emd_37752_validation.pdf.gz | 674.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37752_full_validation.pdf.gz emd_37752_full_validation.pdf.gz | 674.1 KB | Display | |
| Data in XML |  emd_37752_validation.xml.gz emd_37752_validation.xml.gz | 13.1 KB | Display | |
| Data in CIF |  emd_37752_validation.cif.gz emd_37752_validation.cif.gz | 15.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37752 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37752 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37752 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37752 | HTTPS FTP |
-Related structure data
| Related structure data |  8wqrMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37752.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37752.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37752_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
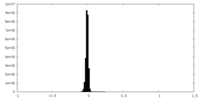
| Density Histograms |
-Half map: #1
| File | emd_37752_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
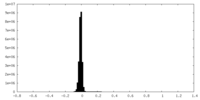
| Density Histograms |
- Sample components
Sample components
-Entire : DDB1-AMBRA1 complex
| Entire | Name: DDB1-AMBRA1 complex |
|---|---|
| Components |
|
-Supramolecule #1: DDB1-AMBRA1 complex
| Supramolecule | Name: DDB1-AMBRA1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Activating molecule in BECN1-regulated autophagy protein 1
| Macromolecule | Name: Activating molecule in BECN1-regulated autophagy protein 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.496871 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKVVPEKNAV RILWGRERGA RAMGAQRLLQ ELVEDKTRWM KWEGKRVELP DSPRSTFLLA FSPDRTLLAS THVNHNIYIT EVKTGKCVH SLIGHRRTPW CVTFHPTISG LIASGCLDGE VRIWDLHGGS ESWFTDSNNA IASLAFHPTA QLLLIATANE I HFWDWSRR ...String: MKVVPEKNAV RILWGRERGA RAMGAQRLLQ ELVEDKTRWM KWEGKRVELP DSPRSTFLLA FSPDRTLLAS THVNHNIYIT EVKTGKCVH SLIGHRRTPW CVTFHPTISG LIASGCLDGE VRIWDLHGGS ESWFTDSNNA IASLAFHPTA QLLLIATANE I HFWDWSRR EPFAVVKTAS EMERVRLVRF DPLGHYLLTA IVNPSNSNIA NTTYRLQWWD FTKFDLPEIS NASVNVLVQN CK IYNDASC DISADGQLLA AFIPSSQRGF PDEGILAVYS LAPHNLGEML YTKRFGPNAI SVSLSPMGRY VMVGLASRRI LLH PSTEHM VAQVFRLQQA HGGETSMRRV FNVLYPMPAD QRRHVSINSA RWLPEPGLGL AYGTNKGDLV ICRPEALNSG UniProtKB: Activating molecule in BECN1-regulated autophagy protein 1, Activating molecule in BECN1-regulated autophagy protein 1 |
-Macromolecule #2: DNA damage-binding protein 1
| Macromolecule | Name: DNA damage-binding protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 127.097469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK ...String: MSYNYVVTAQ KPTAVNGCVT GHFTSAEDLN LLIAKNTRLE IYVVTAEGLR PVKEVGMYGK IAVMELFRPK GESKDLLFIL TAKYNACIL EYKQSGESID IITRAHGNVQ DRIGRPSETG IIGIIDPECR MIGLRLYDGL FKVIPLDRDN KELKAFNIRL E ELHVIDVK FLYGCQAPTI CFVYQDPQGR HVKTYEVSLR EKEFNKGPWK QENVEAEASM VIAVPEPFGG AIIIGQESIT YH NGDKYLA IAPPIIKQST IVCHNRVDPN GSRYLLGDME GRLFMLLLEK EEQMDGTVTL KDLRVELLGE TSIAECLTYL DNG VVFVGS RLGDSQLVKL NVDSNEQGSY VVAMETFTNL GPIVDMCVVD LERQGQGQLV TCSGAFKEGS LRIIRNGIGI HEHA SIDLP GIKGLWPLRS DPNRETDDTL VLSFVGQTRV LMLNGEEVEE TELMGFVDDQ QTFFCGNVAH QQLIQITSAS VRLVS QEPK ALVSEWKEPQ AKNISVASCN SSQVVVAVGR ALYYLQIHPQ ELRQISHTEM EHEVACLDIT PLGDSNGLSP LCAIGL WTD ISARILKLPS FELLHKEMLG GEIIPRSILM TTFESSHYLL CALGDGALFY FGLNIETGLL SDRKKVTLGT QPTVLRT FR SLSTTNVFAC SDRPTVIYSS NHKLVFSNVN LKEVNYMCPL NSDGYPDSLA LANNSTLTIG TIDEIQKLHI RTVPLYES P RKICYQEVSQ CFGVLSSRIE VQDTSGGTTA LRPSASTQAL SSSVSSSKLF SSSTAPHETS FGEEVEVHNL LIIDQHTFE VLHAHQFLQN EYALSLVSCK LGKDPNTYFI VGTAMVYPEE AEPKQGRIVV FQYSDGKLQT VAEKEVKGAV YSMVEFNGKL LASINSTVR LYEWTTEKEL RTECNHYNNI MALYLKTKGD FILVGDLMRS VLLLAYKPME GNFEEIARDF NPNWMSAVEI L DDDNFLGA ENAFNLFVCQ KDSAATTDEE RQHLQEVGLF HLGEFVNVFC HGSLVMQNLG ETSTPTQGSV LFGTVNGMIG LV TSLSESW YNLLLDMQNR LNKVIKSVGK IEHSFWRSFH TERKTEPATG FIDGDLIESF LDISRPKMQE VVANLQYDDG SGM KREATA DDLIKVVEEL TRIH UniProtKB: DNA damage-binding protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Grid information as following: Company/model: Quantifoil Cu 1.2/1.3 Material:Cu Grid mesh: 200 |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 58.96 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)