+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local refinement of FEM1B bound with the C-degron of CUX1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | E3 ubiquitin ligase / Pro/C-degron / PROTEIN BINDING | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of ubiquitin-protein transferase activity / epithelial cell maturation involved in prostate gland development / Intra-Golgi traffic / branching involved in prostate gland morphogenesis / intra-Golgi vesicle-mediated transport / regulation of DNA damage checkpoint / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / death receptor binding / regulation of extrinsic apoptotic signaling pathway via death domain receptors / Cul2-RING ubiquitin ligase complex ...regulation of ubiquitin-protein transferase activity / epithelial cell maturation involved in prostate gland development / Intra-Golgi traffic / branching involved in prostate gland morphogenesis / intra-Golgi vesicle-mediated transport / regulation of DNA damage checkpoint / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / death receptor binding / regulation of extrinsic apoptotic signaling pathway via death domain receptors / Cul2-RING ubiquitin ligase complex / ubiquitin ligase complex / ubiquitin-like ligase-substrate adaptor activity / Neddylation / proteasome-mediated ubiquitin-dependent protein catabolic process / protein ubiquitination / Golgi membrane / apoptotic process / Golgi apparatus / mitochondrion / nucleoplasm / metal ion binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Chen X / Zhang K / Xu C | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Mechanism of Ψ-Pro/C-degron recognition by the CRL2 ubiquitin ligase. Authors: Xinyan Chen / Anat Raiff / Shanshan Li / Qiong Guo / Jiahai Zhang / Hualin Zhou / Richard T Timms / Xuebiao Yao / Stephen J Elledge / Itay Koren / Kaiming Zhang / Chao Xu /     Abstract: The E3 ligase-degron interaction determines the specificity of the ubiquitin‒proteasome system. We recently discovered that FEM1B, a substrate receptor of Cullin 2-RING ligase (CRL2), recognizes C- ...The E3 ligase-degron interaction determines the specificity of the ubiquitin‒proteasome system. We recently discovered that FEM1B, a substrate receptor of Cullin 2-RING ligase (CRL2), recognizes C-degrons containing a C-terminal proline. By solving several cryo-EM structures of CRL2 bound to different C-degrons, we elucidate the dimeric assembly of the complex. Furthermore, we reveal distinct dimerization states of unmodified and neddylated CRL2 to uncover the NEDD8-mediated activation mechanism of CRL2. Our research also indicates that, FEM1B utilizes a bipartite mechanism to recognize both the C-terminal proline and an upstream aromatic residue within the substrate. These structural findings, complemented by in vitro ubiquitination and in vivo cell-based assays, demonstrate that CRL2-mediated polyubiquitination and subsequent protein turnover depend on both FEM1B-degron interactions and the dimerization state of the E3 ligase complex. Overall, this study deepens our molecular understanding of how Cullin-RING E3 ligase substrate selection mediates protein turnover. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37746.map.gz emd_37746.map.gz | 483.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37746-v30.xml emd-37746-v30.xml emd-37746.xml emd-37746.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37746.png emd_37746.png | 46.1 KB | ||
| Filedesc metadata |  emd-37746.cif.gz emd-37746.cif.gz | 5.8 KB | ||
| Others |  emd_37746_half_map_1.map.gz emd_37746_half_map_1.map.gz emd_37746_half_map_2.map.gz emd_37746_half_map_2.map.gz | 474.4 MB 474.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37746 http://ftp.pdbj.org/pub/emdb/structures/EMD-37746 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37746 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37746 | HTTPS FTP |
-Related structure data
| Related structure data |  8wqiMC  8wqaC  8wqbC  8wqcC  8wqdC  8wqeC  8wqfC  8wqgC  8wqhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37746.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37746.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
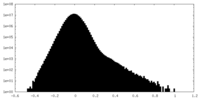
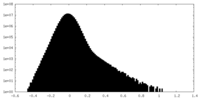
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_37746_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
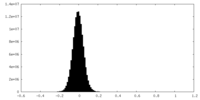
| Density Histograms |
-Half map: #2
| File | emd_37746_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
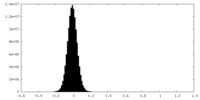
| Density Histograms |
- Sample components
Sample components
-Entire : Local refinement of FEM1B bound with the C-degron of CUX1
| Entire | Name: Local refinement of FEM1B bound with the C-degron of CUX1 |
|---|---|
| Components |
|
-Supramolecule #1: Local refinement of FEM1B bound with the C-degron of CUX1
| Supramolecule | Name: Local refinement of FEM1B bound with the C-degron of CUX1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Protein fem-1 homolog B
| Macromolecule | Name: Protein fem-1 homolog B / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.355062 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEGLAGYVYK AASEGKVLTL AALLLNRSES DIRYLLGYVS QQGGQRSTPL IIAARNGHAK VVRLLLEHYR VQTQQTGTVR FDGYVIDGA TALWCAAGAG HFEVVKLLVS HGANVNHTTV TNSTPLRAAC FDGRLDIVKY LVENNANISI ANKYDNTCLM I AAYKGHTD ...String: MEGLAGYVYK AASEGKVLTL AALLLNRSES DIRYLLGYVS QQGGQRSTPL IIAARNGHAK VVRLLLEHYR VQTQQTGTVR FDGYVIDGA TALWCAAGAG HFEVVKLLVS HGANVNHTTV TNSTPLRAAC FDGRLDIVKY LVENNANISI ANKYDNTCLM I AAYKGHTD VVRYLLEQRA DPNAKAHCGA TALHFAAEAG HIDIVKELIK WRAAIVVNGH GMTPLKVAAE SCKADVVELL LS HADCDRR SRIEALELLG ASFANDRENY DIIKTYHYLY LAMLERFQDG DNILEKEVLP PIHAYGNRTE CRNPQELESI RQD RDALHM EGLIVRERIL GADNIDVSHP IIYRGAVYAD NMEFEQCIKL WLHALHLRQK GNRNTHKDLL RFAQVFSQMI HLNE TVKAP DIECVLRCSV LEIEQSMNRV KNISDADVHN AMDNYECNLY TFLYLVCIST KTQCSEEDQC KINKQIYNLI HLDPR TREG FTLLHLAVNS NTPVDDFHTN DVCSFPNALV TKLLLDCGAE VNAVDNEGNS ALHIIVQYNR PISDFLTLHS IIISLV EAG AHTDMTNKQN KTPLDKSTTG VSEILLKTQM KMSLKCLAAR AVRANDINYQ DQIPRTLEEF VGFH UniProtKB: Protein fem-1 homolog B |
-Macromolecule #2: Protein CASP
| Macromolecule | Name: Protein CASP / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.143281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGGSGGGSKF ADHLHKFHEN DNGAAAGDLW Q UniProtKB: Protein CASP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 8112 / Average electron dose: 57.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 206765 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)