+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo EM map of SLC7A10 in the apo state | |||||||||
 Map data Map data | cryo EM map of SLC7A10 in the apo state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SLC7A10 / SLC3A2 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationD-serine transmembrane transport / positive regulation of synaptic transmission, glycinergic / D-alanine transmembrane transport / glycine transport / L-serine transmembrane transporter activity / negative regulation of brown fat cell differentiation / apical pole of neuron / tyrosine transport / L-histidine transport / amino acid transport complex ...D-serine transmembrane transport / positive regulation of synaptic transmission, glycinergic / D-alanine transmembrane transport / glycine transport / L-serine transmembrane transporter activity / negative regulation of brown fat cell differentiation / apical pole of neuron / tyrosine transport / L-histidine transport / amino acid transport complex / L-leucine import across plasma membrane / L-alanine transmembrane transporter activity / L-alanine import across plasma membrane / Defective SLC7A7 causes lysinuric protein intolerance (LPI) / aromatic amino acid transmembrane transporter activity / phenylalanine transport / methionine transport / L-leucine transmembrane transporter activity / isoleucine transport / valine transport / proline transport / L-amino acid transmembrane transporter activity / L-leucine transport / thyroid hormone transport / neutral amino acid transport / neutral L-amino acid transmembrane transporter activity / Tryptophan catabolism / Amino acid transport across the plasma membrane / exogenous protein binding / anchoring junction / Basigin interactions / response to exogenous dsRNA / amino acid transport / tryptophan transport / basal plasma membrane / calcium ion transport / melanosome / double-stranded RNA binding / virus receptor activity / basolateral plasma membrane / carbohydrate metabolic process / apical plasma membrane / cadherin binding / protein heterodimerization activity / lysosomal membrane / synapse / symbiont entry into host cell / cell surface / protein homodimerization activity / RNA binding / extracellular exosome / nucleoplasm / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.42 Å | |||||||||
 Authors Authors | Li YN / Guo YY / Dai L / Yan RH | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structure of the human Asc-1 transporter complex. Authors: Yaning Li / Yingying Guo / Angelika Bröer / Lu Dai / Stefan Brӧer / Renhong Yan /   Abstract: The Alanine-Serine-Cysteine transporter 1 (Asc-1 or SLC7A10) forms a crucial heterodimeric transporter complex with 4F2hc (SLC3A2) through a covalent disulfide bridge. This complex enables the sodium- ...The Alanine-Serine-Cysteine transporter 1 (Asc-1 or SLC7A10) forms a crucial heterodimeric transporter complex with 4F2hc (SLC3A2) through a covalent disulfide bridge. This complex enables the sodium-independent transport of small neutral amino acids, including L-Alanine (L-Ala), Glycine (Gly), and D-Serine (D-Ser), within the central nervous system (CNS). D-Ser and Gly are two key endogenous glutamate co-agonists that activate N-methyl-d-aspartate (NMDA) receptors by binding to the allosteric site. Mice deficient in Asc-1 display severe symptoms such as tremors, ataxia, and seizures, leading to early postnatal death. Despite its physiological importance, the functional mechanism of the Asc-1-4F2hc complex has remained elusive. Here, we present cryo-electron microscopy (cryo-EM) structures of the human Asc-1-4F2hc complex in its apo state, D-Ser bound state, and L-Ala bound state, resolved at 3.6 Å, 3.5 Å, and 3.4 Å, respectively. Through detailed structural analysis and transport assays, we uncover a comprehensive alternating access mechanism that underlies conformational changes in the complex. In summary, our findings reveal the architecture of the Asc-1 and 4F2hc complex and provide valuable insights into substrate recognition and the functional cycle of this essential transporter complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37671.map.gz emd_37671.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37671-v30.xml emd-37671-v30.xml emd-37671.xml emd-37671.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37671.png emd_37671.png | 49.2 KB | ||
| Filedesc metadata |  emd-37671.cif.gz emd-37671.cif.gz | 6 KB | ||
| Others |  emd_37671_half_map_1.map.gz emd_37671_half_map_1.map.gz emd_37671_half_map_2.map.gz emd_37671_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37671 http://ftp.pdbj.org/pub/emdb/structures/EMD-37671 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37671 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37671 | HTTPS FTP |
-Validation report
| Summary document |  emd_37671_validation.pdf.gz emd_37671_validation.pdf.gz | 705.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37671_full_validation.pdf.gz emd_37671_full_validation.pdf.gz | 704.7 KB | Display | |
| Data in XML |  emd_37671_validation.xml.gz emd_37671_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_37671_validation.cif.gz emd_37671_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37671 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37671 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37671 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37671 | HTTPS FTP |
-Related structure data
| Related structure data |  8wnsMC  8wntC  8wnyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37671.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37671.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo EM map of SLC7A10 in the apo state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||
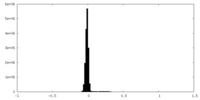
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: cryo EM half map of SLC7A10 in the apo state
| File | emd_37671_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo EM half map of SLC7A10 in the apo state | ||||||||||||
| Projections & Slices |
| ||||||||||||
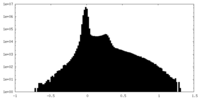
| Density Histograms |
-Half map: cryo EM half map of SLC7A10 in the apo state
| File | emd_37671_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo EM half map of SLC7A10 in the apo state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 4F2hc-ASC-1 complex
| Entire | Name: 4F2hc-ASC-1 complex |
|---|---|
| Components |
|
-Supramolecule #1: 4F2hc-ASC-1 complex
| Supramolecule | Name: 4F2hc-ASC-1 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Amino acid transporter heavy chain SLC3A2
| Macromolecule | Name: Amino acid transporter heavy chain SLC3A2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 68.16468 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MELQPPEASI AVVSIPRQLP GSHSEAGVQG LSAGDDSETG SDCVTQAGLQ LLASSDPPAL ASKNAEVTVE TGFHHVSQAD IEFLTSIDP TASASGSAGI TGTMSQDTEV DMKEVELNEL EPEKQPMNAA SGAAMSLAGA EKNGLVKIKV AEDEAEAAAA A KFTGLSKE ...String: MELQPPEASI AVVSIPRQLP GSHSEAGVQG LSAGDDSETG SDCVTQAGLQ LLASSDPPAL ASKNAEVTVE TGFHHVSQAD IEFLTSIDP TASASGSAGI TGTMSQDTEV DMKEVELNEL EPEKQPMNAA SGAAMSLAGA EKNGLVKIKV AEDEAEAAAA A KFTGLSKE ELLKVAGSPG WVRTRWALLL LFWLGWLGML AGAVVIIVRA PRCRELPAQK WWHTGALYRI GDLQAFQGHG AG NLAGLKG RLDYLSSLKV KGLVLGPIHK NQKDDVAQTD LLQIDPNFGS KEDFDSLLQS AKKKSIRVIL DLTPNYRGEN SWF STQVDT VATKVKDALE FWLQAGVDGF QVRDIENLKD ASSFLAEWQN ITKGFSEDRL LIAGTNSSDL QQILSLLESN KDLL LTSSY LSDSGSTGEH TKSLVTQYLN ATGNRWCSWS LSQARLLTSF LPAQLLRLYQ LMLFTLPGTP VFSYGDEIGL DAAAL PGQP MEAPVMLWDE SSFPDIPGAV SANMTVKGQS EDPGSLLSLF RRLSDQRSKE RSLLHGDFHA FSAGPGLFSY IRHWDQ NER FLVVLNFGDV GLSAGLQASD LPASASLPAK ADLLLSTQPG REEGSPLELE RLKLEPHEGL LLRFPYAA UniProtKB: Amino acid transporter heavy chain SLC3A2 |
-Macromolecule #2: Asc-type amino acid transporter 1
| Macromolecule | Name: Asc-type amino acid transporter 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.837504 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGHTQQPSG RGNPRPAPSP SPVPGTVPGA SERVALKKEI GLLSACTIII GNIIGSGIFI SPKGVLEHSG SVGLALFVWV LGGGVTALG SLCYAELGVA IPKSGGDYAY VTEIFGGLAG FLLLWSAVLI MYPTSLAVIS MTFSNYVLQP VFPNCIPPTT A SRVLSMAC ...String: MAGHTQQPSG RGNPRPAPSP SPVPGTVPGA SERVALKKEI GLLSACTIII GNIIGSGIFI SPKGVLEHSG SVGLALFVWV LGGGVTALG SLCYAELGVA IPKSGGDYAY VTEIFGGLAG FLLLWSAVLI MYPTSLAVIS MTFSNYVLQP VFPNCIPPTT A SRVLSMAC LMLLTWVNSS SVRWATRIQD MFTGGKLLAL SLIIGVGLLQ IFQGHFEELR PSNAFAFWMT PSVGHLALAF LQ GSFAFSG WNFLNYVTEE MVDARKNLPR AIFISIPLVT FVYTFTNIAY FTAMSPQELL SSNAVAVTFG EKLLGYFSWV MPV SVALST FGGINGYLFT YSRLCFSGAR EGHLPSLLAM IHVRHCTPIP ALLVCCGATA VIMLVGDTYT LINYVSFINY LCYG VTILG LLLLRWRRPA LHRPIKVNLL IPVAYLVFWA FLLVFSFISE PMVCGVGVII ILTGVPIFFL GVFWRSKPKC VHRLT ESMT HWGQELCFVV YPQDAPEEEE NGPCPPSLLP ATDKPSKPQ UniProtKB: Asc-type amino acid transporter 1 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.42 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 300000 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)