+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | mGlu2-mGlu4 heterodimer bound mGlu4 agonist E7P | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationadenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway / regulation of response to drug / group II metabotropic glutamate receptor activity / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / intracellular glutamate homeostasis / behavioral response to nicotine / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / neurotransmitter secretion ...adenylate cyclase-inhibiting G protein-coupled glutamate receptor signaling pathway / regulation of response to drug / group II metabotropic glutamate receptor activity / adenylate cyclase inhibiting G protein-coupled glutamate receptor activity / intracellular glutamate homeostasis / behavioral response to nicotine / negative regulation of adenylate cyclase activity / G protein-coupled glutamate receptor signaling pathway / Class C/3 (Metabotropic glutamate/pheromone receptors) / neurotransmitter secretion / glutamate secretion / glutamate receptor activity / long-term synaptic depression / regulation of glutamate secretion / astrocyte projection / cellular response to stress / regulation of dopamine secretion / regulation of synaptic transmission, glutamatergic / presynaptic modulation of chemical synaptic transmission / regulation of neuron apoptotic process / response to cocaine / calcium channel regulator activity / G protein-coupled receptor activity / Sensory perception of sweet, bitter, and umami (glutamate) taste / presynapse / presynaptic membrane / cytoplasmic vesicle / scaffold protein binding / gene expression / G alpha (i) signalling events / chemical synaptic transmission / postsynaptic membrane / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of MAPK cascade / axon / dendrite / glutamatergic synapse / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.46 Å | |||||||||
 Authors Authors | Zhang Y / Liu J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis of orientated asymmetry in a mGlu heterodimer. Authors: Weizhu Huang / Nan Jin / Jia Guo / Cangsong Shen / Chanjuan Xu / Kun Xi / Léo Bonhomme / Robert B Quast / Dan-Dan Shen / Jiao Qin / Yi-Ru Liu / Yuxuan Song / Yang Gao / Emmanuel Margeat / ...Authors: Weizhu Huang / Nan Jin / Jia Guo / Cangsong Shen / Chanjuan Xu / Kun Xi / Léo Bonhomme / Robert B Quast / Dan-Dan Shen / Jiao Qin / Yi-Ru Liu / Yuxuan Song / Yang Gao / Emmanuel Margeat / Philippe Rondard / Jean-Philippe Pin / Yan Zhang / Jianfeng Liu /   Abstract: The structural basis for the allosteric interactions within G protein-coupled receptors (GPCRs) heterodimers remains largely unknown. The metabotropic glutamate (mGlu) receptors are complex dimeric ...The structural basis for the allosteric interactions within G protein-coupled receptors (GPCRs) heterodimers remains largely unknown. The metabotropic glutamate (mGlu) receptors are complex dimeric GPCRs important for the fine tuning of many synapses. Heterodimeric mGlu receptors with specific allosteric properties have been identified in the brain. Here we report four cryo-electron microscopy structures of mGlu2-4 heterodimer in different states: an inactive state bound to antagonists, two intermediate states bound to either mGlu2 or mGlu4 agonist only and an active state bound to both glutamate and a mGlu4 positive allosteric modulator (PAM) in complex with Gi protein. In addition to revealing a unique PAM binding pocket among mGlu receptors, our data bring important information for the asymmetric activation of mGlu heterodimers. First, we show that agonist binding to a single subunit in the extracellular domain is not sufficient to stabilize an active dimer conformation. Single-molecule FRET data show that the monoliganded mGlu2-4 can be found in both intermediate states and an active one. Second, we provide a detailed view of the asymmetric interface in seven-transmembrane (7TM) domains and identified key residues within the mGlu2 7TM that limits its activation leaving mGlu4 as the only subunit activating G proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37506.map.gz emd_37506.map.gz | 258.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37506-v30.xml emd-37506-v30.xml emd-37506.xml emd-37506.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37506.png emd_37506.png | 40.3 KB | ||
| Filedesc metadata |  emd-37506.cif.gz emd-37506.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37506 http://ftp.pdbj.org/pub/emdb/structures/EMD-37506 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37506 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37506 | HTTPS FTP |
-Related structure data
| Related structure data |  8wg9MC  8wgbC  8wgcC  8wgdC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37506.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37506.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : mGlu2-mGlu4 heterodimer bound L-AP4
| Entire | Name: mGlu2-mGlu4 heterodimer bound L-AP4 |
|---|---|
| Components |
|
-Supramolecule #1: mGlu2-mGlu4 heterodimer bound L-AP4
| Supramolecule | Name: mGlu2-mGlu4 heterodimer bound L-AP4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Metabotropic glutamate receptor 4
| Macromolecule | Name: Metabotropic glutamate receptor 4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.36532 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KPKGHPHMNS IRIDGDITLG GLFPVHGRGS EGKPCGELKK EKGIHRLEAM LFALDRINND PDLLPNITLG ARILDTCSRD THALEQSLT FVQALIEKDG TEVRCGSGGP PIITKPERVV GVIGASGSSV SIMVANILRL FKIPQISYAS TAPDLSDNSR Y DFFSRVVP ...String: KPKGHPHMNS IRIDGDITLG GLFPVHGRGS EGKPCGELKK EKGIHRLEAM LFALDRINND PDLLPNITLG ARILDTCSRD THALEQSLT FVQALIEKDG TEVRCGSGGP PIITKPERVV GVIGASGSSV SIMVANILRL FKIPQISYAS TAPDLSDNSR Y DFFSRVVP SDTYQAQAMV DIVRALKWNY VSTVASEGSY GESGVEAFIQ KSREDGGVCI AQSVKIPREP KAGEFDKIIR RL LETSNAR AVIIFANEDD IRRVLEAARR ANQTGHFFWM GSDSWGSKIA PVLHLEEVAE GAVTILPKRM SVRGFDRYFS SRT LDNNRR NIWFAEFWED NFHCKLSRHA LKKGSHVKKC TNRERIGQDS AYEQEGKVQF VIDAVYAMGH ALHAMHRDLC PGRV GLCPR MDPVDGTQLL KYIRNVNFSG IAGNPVTFNE NGDAPGRYDI YQYQLRNDSA EYKVIGSWTD HLHLRIERMH WPGSG QQLP RSICSLPCQP GERKKTVKGM PCCWHCEPCT GYQYQVDRYT CKTCPYDMRP TENRTGCRPI PIIKLEWGSP WAVLPL FLA VVGIAATLFV VITFVRYNDT PIVKASGREL SYVLLAGIFL CYATTFLMIA EPDLGTCSLR RIFLGLGMSI SYAALLT KT NRIYRIFEQG KRSVSAPRFI SPASQLAITF SLISLQLLGI CVWFVVDPSH SVVDFQDQRT LDPRFARGVL KCDISDLS L ICLLGYSMLL MVTCTVYAIK TRGVPETFNE AKPIGFTMYT TCIVWLAFIP IFFGTSQSAD KLYIQTTTLT VSVSLSASV SLGMLYMPKV YIILFHPEQN VPKRKRSLKA VVTAATMSNK FTQKGNFRPN GEAKSELCEN LEAPALATKQ TYVTYTNHAI UniProtKB: Metabotropic glutamate receptor 4 |
-Macromolecule #2: Metabotropic glutamate receptor 2
| Macromolecule | Name: Metabotropic glutamate receptor 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.861367 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EGPAKKVLTL EGDLVLGGLF PVHQKGGPAE DCGPVNEHRG IQRLEAMLFA LDRINRDPHL LPGVRLGAHI LDSCSKDTHA LEQALDFVR ASLSRGADGS RHICPDGSYA THGDAPTAIT GVIGGSYSDV SIQVANLLRL FQIPQISYAS TSAKLSDKSR Y DYFARTVP ...String: EGPAKKVLTL EGDLVLGGLF PVHQKGGPAE DCGPVNEHRG IQRLEAMLFA LDRINRDPHL LPGVRLGAHI LDSCSKDTHA LEQALDFVR ASLSRGADGS RHICPDGSYA THGDAPTAIT GVIGGSYSDV SIQVANLLRL FQIPQISYAS TSAKLSDKSR Y DYFARTVP PDFFQAKAMA EILRFFNWTY VSTVASEGDY GETGIEAFEL EARARNICVA TSEKVGRAMS RAAFEGVVRA LL QKPSARV AVLFTRSEDA RELLAASQRL NASFTWVASD GWGALESVVA GSEGAAEGAI TIELASYPIS DFASYFQSLD PWN NSRNPW FREFWEQRFR CSFRQRDCAA HSLRAVPFEQ ESKIMFVVNA VYAMAHALHN MHRALCPNTT RLCDAMRPVN GRRL YKDFV LNVKFDAPFR PADTHNEVRF DRFGDGIGRY NIFTYLRAGS GRYRYQKVGY WAEGLTLDTS LIPWASPSAG PLPAS RCSE PCLQNEVKSV QPGEVCCWLC IPCQPYEYRL DEFTCADCGL GYWPNASLTG CFELPQEYIR WGDAWAVGPV TIACLG ALA TLFVLGVFVR HNATPVVKAS GRELCYILLG GVFLCYCMTF IFIAKPSTAV CTLRRLGLGT AFSVCYSALL TKTNRIA RI FGGAREGAQR PRFISPASQV AICLALISGQ LLIVVAWLVV EAPGTGKETA PERREVVTLR CNHRDASMLG SLAYNVLL I ALCTLYAFKT RKCPENFNEA KFIGFTMYTT CIIWLAFLPI FYVTSSDYRV QTTTMCVSVS LSGSVVLGCL FAPKLHIIL FQPQKNVVSH RAPTSRFGSA AARASSSLGQ GSGSQFVPTV CNGREVVDST TSSL UniProtKB: Metabotropic glutamate receptor 2 |
-Macromolecule #3: (2S)-2-amino-4-phosphonobutanoic acid
| Macromolecule | Name: (2S)-2-amino-4-phosphonobutanoic acid / type: ligand / ID: 3 / Number of copies: 1 / Formula: E7P |
|---|---|
| Molecular weight | Theoretical: 183.1 Da |
| Chemical component information | 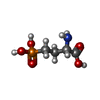 ChemComp-E7P: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















