[English] 日本語
 Yorodumi
Yorodumi- EMDB-37492: human glycine transporter 1 in complex with glycine in occluded c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human glycine transporter 1 in complex with glycine in occluded conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GlyT1 / glycine / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of synaptic transmission, glycinergic / glycine secretion, neurotransmission / glycine transmembrane transporter activity / glycine:sodium symporter activity / positive regulation of heme biosynthetic process / neurotransmitter uptake / glycine import across plasma membrane / glycine transport / positive regulation of hemoglobin biosynthetic process / amino acid:sodium symporter activity ...regulation of synaptic transmission, glycinergic / glycine secretion, neurotransmission / glycine transmembrane transporter activity / glycine:sodium symporter activity / positive regulation of heme biosynthetic process / neurotransmitter uptake / glycine import across plasma membrane / glycine transport / positive regulation of hemoglobin biosynthetic process / amino acid:sodium symporter activity / dense core granule / SLC-mediated transport of neurotransmitters / parallel fiber to Purkinje cell synapse / lateral plasma membrane / transport across blood-brain barrier / sodium ion transmembrane transport / basal plasma membrane / hippocampal mossy fiber to CA3 synapse / synaptic vesicle membrane / presynaptic membrane / basolateral plasma membrane / postsynaptic membrane / endosome / postsynaptic density / apical plasma membrane / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.58 Å | |||||||||
 Authors Authors | Wei Y / Zhao Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Transport mechanism and pharmacology of the human GlyT1. Authors: Yiqing Wei / Renjie Li / Yufei Meng / Tuo Hu / Jun Zhao / Yiwei Gao / Qinru Bai / Na Li / Yan Zhao /  Abstract: The glycine transporter 1 (GlyT1) plays a crucial role in the regulation of both inhibitory and excitatory neurotransmission by removing glycine from the synaptic cleft. Given its close association ...The glycine transporter 1 (GlyT1) plays a crucial role in the regulation of both inhibitory and excitatory neurotransmission by removing glycine from the synaptic cleft. Given its close association with glutamate/glycine co-activated NMDA receptors (NMDARs), GlyT1 has emerged as a central target for the treatment of schizophrenia, which is often linked to hypofunctional NMDARs. Here, we report the cryo-EM structures of GlyT1 bound with substrate glycine and drugs ALX-5407, SSR504734, and PF-03463275. These structures, captured at three fundamental states of the transport cycle-outward-facing, occluded, and inward-facing-enable us to illustrate a comprehensive blueprint of the conformational change associated with glycine reuptake. Additionally, we identified three specific pockets accommodating drugs, providing clear insights into the structural basis of their inhibitory mechanism and selectivity. Collectively, these structures offer significant insights into the transport mechanism and recognition of substrate and anti-schizophrenia drugs, thus providing a platform to design small molecules to treat schizophrenia. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37492.map.gz emd_37492.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37492-v30.xml emd-37492-v30.xml emd-37492.xml emd-37492.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37492.png emd_37492.png | 40.3 KB | ||
| Filedesc metadata |  emd-37492.cif.gz emd-37492.cif.gz | 6.1 KB | ||
| Others |  emd_37492_half_map_1.map.gz emd_37492_half_map_1.map.gz emd_37492_half_map_2.map.gz emd_37492_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37492 http://ftp.pdbj.org/pub/emdb/structures/EMD-37492 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37492 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37492 | HTTPS FTP |
-Related structure data
| Related structure data |  8wfiMC  8wfjC  8wfkC  8wflC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37492.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37492.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
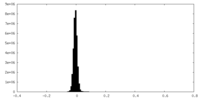
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37492_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
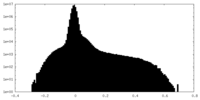
| Density Histograms |
-Half map: #1
| File | emd_37492_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
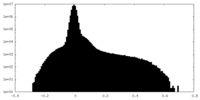
| Density Histograms |
- Sample components
Sample components
-Entire : human glycine transporter 1 (GlyT1)
| Entire | Name: human glycine transporter 1 (GlyT1) |
|---|---|
| Components |
|
-Supramolecule #1: human glycine transporter 1 (GlyT1)
| Supramolecule | Name: human glycine transporter 1 (GlyT1) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform GlyT-1B of Sodium- and chloride-dependent glycine transpo...
| Macromolecule | Name: Isoform GlyT-1B of Sodium- and chloride-dependent glycine transporter 1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.769484 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RGNWGNQIEF VLTSVGYAVG LGNVWRFPYL CYRNAGGAFM FPYFIMLIFC GIPLFFMELS FGQFASQGCL GVWRISPMFK GVGYGMMVV STYIGIYYNV VICIAFYYFF SSMTHVLPWA YCNNPWNTHD CAGVLDASNL TNGSRPAALP SNLSHLLNHS L QRTSPSEE ...String: RGNWGNQIEF VLTSVGYAVG LGNVWRFPYL CYRNAGGAFM FPYFIMLIFC GIPLFFMELS FGQFASQGCL GVWRISPMFK GVGYGMMVV STYIGIYYNV VICIAFYYFF SSMTHVLPWA YCNNPWNTHD CAGVLDASNL TNGSRPAALP SNLSHLLNHS L QRTSPSEE YWRLYVLKLS DDIGNFGEVR LPLLGCLGVS WLVVFLCLIR GVKSSGKVVY FTATFPYVVL TILFVRGVTL EG AFDGIMY YLTPQWDKIL AAKVWGDAAS QIFYSLGCAW GGLITMASYN KFHNNCYRDS VIISITNCAT SVYAGFVIFS ILG FMANHL GVDVSRVADH GPGLAFVAYP EALTLLPISP LWSLLFFFML ILLGLGTQFC LLETLVTAIV DEVGNEWILQ KKTY VTLGV AVAGFLLGIP LTSQAGIYWL LLMDNYAASF SLVVISCIMC VAIMYIYGHR NYFQDIQMML GFPPPLFFQI CWRFV SPAI IFFILVFTVI QYPITAYNHY QYPGWAVAIG FLMALSSVLC IPLYAMFRLC RTDGADLLQR LKNATKPSRD WGPALL EHR TGRYAP UniProtKB: Sodium- and chloride-dependent glycine transporter 1 |
-Macromolecule #2: GLYCINE
| Macromolecule | Name: GLYCINE / type: ligand / ID: 2 / Number of copies: 1 / Formula: GLY |
|---|---|
| Molecular weight | Theoretical: 75.067 Da |
| Chemical component information |  ChemComp-GLY: |
-Macromolecule #3: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 37 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)