+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PSI-LHCI of the red alga Cyanidium caldarium RK-1 (NIES-2137) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Photosystem I / ELECTRON TRANSPORT / PHOTOSYNTHESIS | |||||||||
| Biological species |  Cyanidium caldarium (eukaryote) Cyanidium caldarium (eukaryote) | |||||||||
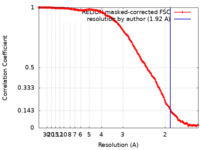
| Method | single particle reconstruction / cryo EM / Resolution: 1.92 Å | |||||||||
 Authors Authors | Kato K / Hamaguchi T / Nakajima Y / Kawakami K / Yonekura K / Shen JR / Nagao R | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: The structure of PSI-LHCI from provides evolutionary insights into conservation and diversity of red-lineage LHCs. Authors: Koji Kato / Tasuku Hamaguchi / Minoru Kumazawa / Yoshiki Nakajima / Kentaro Ifuku / Shunsuke Hirooka / Yuu Hirose / Shin-Ya Miyagishima / Takehiro Suzuki / Keisuke Kawakami / Naoshi Dohmae / ...Authors: Koji Kato / Tasuku Hamaguchi / Minoru Kumazawa / Yoshiki Nakajima / Kentaro Ifuku / Shunsuke Hirooka / Yuu Hirose / Shin-Ya Miyagishima / Takehiro Suzuki / Keisuke Kawakami / Naoshi Dohmae / Koji Yonekura / Jian-Ren Shen / Ryo Nagao /  Abstract: Light-harvesting complexes (LHCs) are diversified among photosynthetic organisms, and the structure of the photosystem I-LHC (PSI-LHCI) supercomplex has been shown to be variable depending on the ...Light-harvesting complexes (LHCs) are diversified among photosynthetic organisms, and the structure of the photosystem I-LHC (PSI-LHCI) supercomplex has been shown to be variable depending on the species of organisms. However, the structural and evolutionary correlations of red-lineage LHCs are unknown. Here, we determined a 1.92-Å resolution cryoelectron microscopic structure of a PSI-LHCI supercomplex isolated from the red alga RK-1 (NIES-2137), which is an important taxon in the Cyanidiophyceae. We subsequently investigated the correlations of PSI-LHCIs from different organisms through structural comparisons and phylogenetic analysis. The PSI-LHCI structure obtained shows five LHCI subunits surrounding a PSI-monomer core. The five LHCIs are composed of two Lhcr1s, two Lhcr2s, and one Lhcr3. Phylogenetic analysis of LHCs bound to PSI in the red-lineage algae showed clear orthology of LHCs between and , whereas no orthologous relationships were found between Lhcr1-3 and LHCs in other red-lineage PSI-LHCI structures. These findings provide evolutionary insights into conservation and diversity of red-lineage LHCs associated with PSI. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37480.map.gz emd_37480.map.gz | 18 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37480-v30.xml emd-37480-v30.xml emd-37480.xml emd-37480.xml | 35.6 KB 35.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37480_fsc.xml emd_37480_fsc.xml | 14 KB | Display |  FSC data file FSC data file |
| Images |  emd_37480.png emd_37480.png | 110.9 KB | ||
| Filedesc metadata |  emd-37480.cif.gz emd-37480.cif.gz | 9.1 KB | ||
| Others |  emd_37480_additional_1.map.gz emd_37480_additional_1.map.gz emd_37480_half_map_1.map.gz emd_37480_half_map_1.map.gz emd_37480_half_map_2.map.gz emd_37480_half_map_2.map.gz | 52.2 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37480 http://ftp.pdbj.org/pub/emdb/structures/EMD-37480 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37480 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37480 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37480.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37480.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.792 Å | ||||||||||||||||||||||||||||||||||||
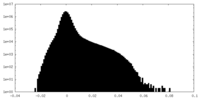
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_37480_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
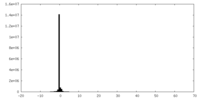
| Density Histograms |
-Half map: #2
| File | emd_37480_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
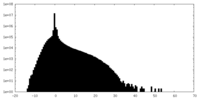
| Density Histograms |
-Half map: #1
| File | emd_37480_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
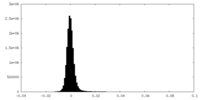
| Density Histograms |
- Sample components
Sample components
+Entire : PSI-LHCI
+Supramolecule #1: PSI-LHCI
+Macromolecule #1: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #2: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #3: Photosystem I iron-sulfur center
+Macromolecule #4: Photosystem I reaction center subunit II
+Macromolecule #5: Photosystem I reaction center subunit IV
+Macromolecule #6: Photosystem I reaction center subunit III
+Macromolecule #7: Photosystem I reaction center subunit VIII
+Macromolecule #8: Photosystem I reaction center subunit IX
+Macromolecule #9: Photosystem I reaction center subunit X
+Macromolecule #10: Photosystem I reaction center subunit XI
+Macromolecule #11: Photosystem I reaction center subunit XII
+Macromolecule #12: Photosystem I subunit O
+Macromolecule #13: Lhcr1
+Macromolecule #14: Lhcr2
+Macromolecule #15: Lhcr3
+Macromolecule #16: CHLOROPHYLL A ISOMER
+Macromolecule #17: CHLOROPHYLL A
+Macromolecule #18: PHYLLOQUINONE
+Macromolecule #19: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #20: BETA-CAROTENE
+Macromolecule #21: IRON/SULFUR CLUSTER
+Macromolecule #22: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #23: UNKNOWN LIGAND
+Macromolecule #24: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #25: Zeaxanthin
+Macromolecule #26: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.83 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| |||||||||
| Grid | Model: Quantifoil R0.6/1 / Mesh: 200 | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Average electron dose: 39.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: in silico model / Details: homology model |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8wey: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)