+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the ABCG25 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | exporter / complex / transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationintercellular transport / negative regulation of post-embryonic development / abscisic acid transport / export from cell / response to abscisic acid / abscisic acid-activated signaling pathway / efflux transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / response to cold ...intercellular transport / negative regulation of post-embryonic development / abscisic acid transport / export from cell / response to abscisic acid / abscisic acid-activated signaling pathway / efflux transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / response to cold / transmembrane transport / response to heat / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
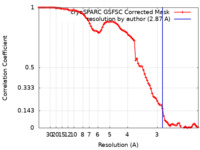
| Method | single particle reconstruction / cryo EM / Resolution: 2.87 Å | |||||||||
 Authors Authors | Xin J / Yan KG | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Plant Commun / Year: 2024 Journal: Plant Commun / Year: 2024Title: Structural insights into AtABCG25, an angiosperm-specific abscisic acid exporter. Authors: Jian Xin / Yeling Zhou / Yichun Qiu / He Geng / Yuzhu Wang / Yi Song / Jiansheng Liang / Kaige Yan /   Abstract: Cellular hormone homeostasis is essential for precise spatial and temporal signaling responses and plant fitness. Abscisic acid (ABA) plays pivotal roles in orchestrating various developmental and ...Cellular hormone homeostasis is essential for precise spatial and temporal signaling responses and plant fitness. Abscisic acid (ABA) plays pivotal roles in orchestrating various developmental and stress responses and confers fitness benefits over ecological and evolutionary timescales in terrestrial plants. Cellular ABA level is regulated by complex processes, including biosynthesis, catabolism, and transport. AtABCG25 is the first ABA exporter identified through genetic screening and affects diverse ABA responses. Resolving the structural basis of ABA export by ABCG25 is critical for further manipulations of ABA homeostasis and plant fitness. We used cryo-electron microscopy to elucidate the structural dynamics of AtABCG25 and successfully characterized different states, including apo AtABCG25, ABA-bound AtABCG25, and ATP-bound AtABCG25 (E232Q). Notably, AtABCG25 forms a homodimer that features a deep, slit-like cavity in the transmembrane domain, and we precisely characterized the critical residues in the cavity where ABA binds. ATP binding triggers closure of the nucleotide-binding domains and conformational transitions in the transmembrane domains. We show that AtABCG25 belongs to a conserved ABCG subfamily that originated during the evolution of angiosperms. This subfamily neofunctionalized to regulate seed germination via the endosperm, in concert with the evolution of this angiosperm-specific, embryo-nourishing tissue. Collectively, these findings provide valuable insights into the intricate substrate recognition and transport mechanisms of the ABA exporter AtABCG25, paving the way for genetic manipulation of ABA homeostasis and plant fitness. #1:  Journal: Acta Crystallogr., Sect. D: Biol. Crystallogr. / Year: 2018 Journal: Acta Crystallogr., Sect. D: Biol. Crystallogr. / Year: 2018Title: Real-space refinement in PHENIX for cryo-EM and crystallography Authors: Xin J / Yan KG | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37455.map.gz emd_37455.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37455-v30.xml emd-37455-v30.xml emd-37455.xml emd-37455.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37455_fsc.xml emd_37455_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_37455.png emd_37455.png | 63.7 KB | ||
| Masks |  emd_37455_msk_1.map emd_37455_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37455.cif.gz emd-37455.cif.gz | 5.6 KB | ||
| Others |  emd_37455_half_map_1.map.gz emd_37455_half_map_1.map.gz emd_37455_half_map_2.map.gz emd_37455_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37455 http://ftp.pdbj.org/pub/emdb/structures/EMD-37455 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37455 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37455 | HTTPS FTP |
-Related structure data
| Related structure data |  8wd6MC  8wamC  8wbaC  8wbxC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37455.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37455.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.095 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37455_msk_1.map emd_37455_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37455_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37455_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the ABCG25
| Entire | Name: Cryo-EM structure of the ABCG25 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the ABCG25
| Supramolecule | Name: Cryo-EM structure of the ABCG25 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ABC transporter G family member 25
| Macromolecule | Name: ABC transporter G family member 25 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 77.121148 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDHDGDY KDHDIDYKDD DDKGSDLEVL FQGPGSMSAF DGVENQMNGP DSSPRLSQDP REPRSLLSSS CFPITLKFVD VCYRVKIHG MSNDSCNIKK LLGLKQKPSD ETRSTEERTI LSGVTGMISP GEFMAVLGPS GSGKSTLLNA VAGRLHGSNL T GKILINDG ...String: MDYKDHDGDY KDHDIDYKDD DDKGSDLEVL FQGPGSMSAF DGVENQMNGP DSSPRLSQDP REPRSLLSSS CFPITLKFVD VCYRVKIHG MSNDSCNIKK LLGLKQKPSD ETRSTEERTI LSGVTGMISP GEFMAVLGPS GSGKSTLLNA VAGRLHGSNL T GKILINDG KITKQTLKRT GFVAQDDLLY PHLTVRETLV FVALLRLPRS LTRDVKLRAA ESVISELGLT KCENTVVGNT FI RGISGGE RKRVSIAHEL LINPSLLVLD EPTSGLDATA ALRLVQTLAG LAHGKGKTVV TSIHQPSSRV FQMFDTVLLL SEG KCLFVG KGRDAMAYFE SVGFSPAFPM NPADFLLDLA NGVCQTDGVT EREKPNVRQT LVTAYDTLLA PQVKTCIEVS HFPQ DNARF VKTRVNGGGI TTCIATWFSQ LCILLHRLLK ERRHESFDLL RIFQVVAASI LCGLMWWHSD YRDVHDRLGL LFFIS IFWG VLPSFNAVFT FPQERAIFTR ERASGMYTLS SYFMAHVLGS LSMELVLPAS FLTFTYWMVY LRPGIVPFLL TLSVLL LYV LASQGLGLAL GAAIMDAKKA STIVTVTMLA FVLTGGYYVN KVPSGMVWMK YVSTTFYCYR LLVAIQYGSG EEILRML GC DSKGKQGASA ATSAGCRFVE EEVIGDVGMW TSVGVLFLMF FGYRVLAYLA LRRIKH UniProtKB: ABC transporter G family member 25 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.5625 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)