[English] 日本語
 Yorodumi
Yorodumi- EMDB-37424: CryoEM structure of non-structural protein 1 tetramer from ZIKA virus -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of non-structural protein 1 tetramer from ZIKA virus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | flavivirus / non-structural protein 1 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationribonucleoside triphosphate phosphatase activity / viral capsid / double-stranded RNA binding / methyltransferase cap1 activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / protein dimerization activity / symbiont-mediated suppression of host innate immune response / host cell endoplasmic reticulum membrane / serine-type endopeptidase activity ...ribonucleoside triphosphate phosphatase activity / viral capsid / double-stranded RNA binding / methyltransferase cap1 activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / protein dimerization activity / symbiont-mediated suppression of host innate immune response / host cell endoplasmic reticulum membrane / serine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / fusion of virus membrane with host endosome membrane / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / virion membrane / structural molecule activity / proteolysis / extracellular region / ATP binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  dengue virus type 4 dengue virus type 4 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Jiao HZ / Pan Q / Hu HL | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: The step-by-step assembly mechanism of secreted flavivirus NS1 tetramer and hexamer captured at atomic resolution. Authors: Qi Pan / Haizhan Jiao / Wanqin Zhang / Qiang Chen / Geshu Zhang / Jianhai Yu / Wei Zhao / Hongli Hu /  Abstract: Flaviviruses encode a conserved, membrane-associated nonstructural protein 1 (NS1) with replication and immune evasion functions. The current knowledge of secreted NS1 (sNS1) oligomers is based on ...Flaviviruses encode a conserved, membrane-associated nonstructural protein 1 (NS1) with replication and immune evasion functions. The current knowledge of secreted NS1 (sNS1) oligomers is based on several low-resolution structures, thus hindering the development of drugs and vaccines against flaviviruses. Here, we revealed that recombinant sNS1 from flaviviruses exists in a dynamic equilibrium of dimer-tetramer-hexamer states. Two DENV4 hexameric NS1 structures and several tetrameric NS1 structures from multiple flaviviruses were solved at atomic resolution by cryo-EM. The stacking of the tetrameric NS1 and hexameric NS1 is facilitated by the hydrophobic β-roll and connector domains. Additionally, a triacylglycerol molecule located within the central cavity may play a role in stabilizing the hexamer. Based on differentiated interactions between the dimeric NS1, two distinct hexamer models (head-to-head and side-to-side hexamer) and the step-by-step assembly mechanisms of NS1 dimer into hexamer were proposed. We believe that our study sheds light on the understanding of the NS1 oligomerization and contributes to NS1-based therapies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37424.map.gz emd_37424.map.gz | 79 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37424-v30.xml emd-37424-v30.xml emd-37424.xml emd-37424.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37424_fsc.xml emd_37424_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_37424.png emd_37424.png | 153.3 KB | ||
| Filedesc metadata |  emd-37424.cif.gz emd-37424.cif.gz | 5.4 KB | ||
| Others |  emd_37424_additional_1.map.gz emd_37424_additional_1.map.gz emd_37424_half_map_1.map.gz emd_37424_half_map_1.map.gz emd_37424_half_map_2.map.gz emd_37424_half_map_2.map.gz | 42 MB 77.7 MB 77.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37424 http://ftp.pdbj.org/pub/emdb/structures/EMD-37424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37424 | HTTPS FTP |
-Validation report
| Summary document |  emd_37424_validation.pdf.gz emd_37424_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37424_full_validation.pdf.gz emd_37424_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_37424_validation.xml.gz emd_37424_validation.xml.gz | 17.5 KB | Display | |
| Data in CIF |  emd_37424_validation.cif.gz emd_37424_validation.cif.gz | 22.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37424 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37424 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37424 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37424 | HTTPS FTP |
-Related structure data
| Related structure data |  8wbgMC  8wbbC  8wbcC  8wbdC  8wbeC  8wbfC  8wbhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37424.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37424.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
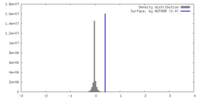
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_37424_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
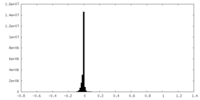
| Density Histograms |
-Half map: #2
| File | emd_37424_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
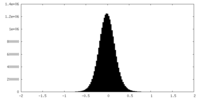
| Density Histograms |
-Half map: #1
| File | emd_37424_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
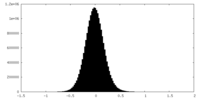
| Density Histograms |
- Sample components
Sample components
-Entire : ZIKA virus non-structural protein 1 tetramer
| Entire | Name: ZIKA virus non-structural protein 1 tetramer |
|---|---|
| Components |
|
-Supramolecule #1: ZIKA virus non-structural protein 1 tetramer
| Supramolecule | Name: ZIKA virus non-structural protein 1 tetramer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  dengue virus type 4 dengue virus type 4 |
-Macromolecule #1: Genome polyprotein
| Macromolecule | Name: Genome polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  dengue virus type 4 dengue virus type 4 |
| Molecular weight | Theoretical: 44.49816 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHVGCS VDFSKKETRC GTGVFVYNDV EAWRDRYKYH PDSPRRLAAA VKQAWEDGIC GISSVSRMEN IMWRSVEGEL NAILEENGV QLTVVVGSVK NPMWRGPQRL PVPVNELPHG WKAWGKSYFV RAAKTNNSFV VDGDTLKECP LKHRAWNSFL V EDHGFGVF ...String: HHHHHHVGCS VDFSKKETRC GTGVFVYNDV EAWRDRYKYH PDSPRRLAAA VKQAWEDGIC GISSVSRMEN IMWRSVEGEL NAILEENGV QLTVVVGSVK NPMWRGPQRL PVPVNELPHG WKAWGKSYFV RAAKTNNSFV VDGDTLKECP LKHRAWNSFL V EDHGFGVF HTSVWLKVRE DYSLECDPAV IGTAVKGKEA VHSDLGYWIE SEKNDTWRLK RAHLIEMKTC EWPKSHTLWT DG IEESDLI IPKSLAGPLS HHNTREGYRT QMKGPWHSEE LEIRFEECPG TKVHVEETCG TRGPSLRSTT ASGRVIEEWC CRE CTMPPL SFRAKDGCWY GMEIRPRKEP ESNLVRSMVT AGSLVPRGSW SHPQFEKGGG SGGGSGGSAW SHPQFEK UniProtKB: Genome polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)