+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the GPR101-Gs complex | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | GPCR / orphan receptor / GPR101 / constitutive activity / cryo-EM / structural protein / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationPKA activation in glucagon signalling / developmental growth / hair follicle placode formation / intracellular transport / D1 dopamine receptor binding / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway ...PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / intracellular transport / D1 dopamine receptor binding / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / electron transport chain / bone development / G protein-coupled receptor activity / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / sensory perception of smell / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / electron transfer activity / periplasmic space / Extra-nuclear estrogen signaling / receptor complex / cell population proliferation / positive regulation of MAPK cascade / iron ion binding / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / heme binding / synapse / GTP binding / protein-containing complex binding / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
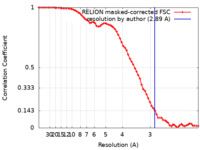
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | ||||||||||||||||||
 Authors Authors | Sun JP / Gao N / Yu X / Wang GP / Yang F / Wang JY / Yang Z / Guan Y | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Structure of GPR101-Gs enables identification of ligands with rejuvenating potential. Authors: Zhao Yang / Jun-Yan Wang / Fan Yang / Kong-Kai Zhu / Guo-Peng Wang / Ying Guan / Shang-Lei Ning / Yan Lu / Yu Li / Chao Zhang / Yuan Zheng / Shu-Hua Zhou / Xin-Wen Wang / Ming-Wei Wang / ...Authors: Zhao Yang / Jun-Yan Wang / Fan Yang / Kong-Kai Zhu / Guo-Peng Wang / Ying Guan / Shang-Lei Ning / Yan Lu / Yu Li / Chao Zhang / Yuan Zheng / Shu-Hua Zhou / Xin-Wen Wang / Ming-Wei Wang / Peng Xiao / Fan Yi / Cheng Zhang / Peng-Ju Zhang / Fei Xu / Bao-Hua Liu / Hua Zhang / Xiao Yu / Ning Gao / Jin-Peng Sun /  Abstract: GPR101 is an orphan G protein-coupled receptor actively participating in energy homeostasis. Here we report the cryo-electron microscopy structure of GPR101 constitutively coupled to Gs heterotrimer, ...GPR101 is an orphan G protein-coupled receptor actively participating in energy homeostasis. Here we report the cryo-electron microscopy structure of GPR101 constitutively coupled to Gs heterotrimer, which reveals unique features of GPR101, including the interaction of extracellular loop 2 within the 7TM bundle, a hydrophobic chain packing-mediated activation mechanism and the structural basis of disease-related mutants. Importantly, a side pocket is identified in GPR101 that facilitates in silico screening to identify four small-molecule agonists, including AA-14. The structure of AA-14-GPR101-Gs provides direct evidence of the AA-14 binding at the side pocket. Functionally, AA-14 partially restores the functions of GH/IGF-1 axis and exhibits several rejuvenating effects in wild-type mice, which are abrogated in Gpr101-deficient mice. In summary, we provide a structural basis for the constitutive activity of GPR101. The structure-facilitated identification of GPR101 agonists and functional analysis suggest that targeting this orphan receptor has rejuvenating potential. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37356.map.gz emd_37356.map.gz | 4.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37356-v30.xml emd-37356-v30.xml emd-37356.xml emd-37356.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37356_fsc.xml emd_37356_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_37356.png emd_37356.png | 34.1 KB | ||
| Filedesc metadata |  emd-37356.cif.gz emd-37356.cif.gz | 6.7 KB | ||
| Others |  emd_37356_half_map_1.map.gz emd_37356_half_map_1.map.gz emd_37356_half_map_2.map.gz emd_37356_half_map_2.map.gz | 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37356 http://ftp.pdbj.org/pub/emdb/structures/EMD-37356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37356 | HTTPS FTP |
-Related structure data
| Related structure data |  8w8qMC  8w8rC  8w8sC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37356.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37356.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.052 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37356_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37356_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the GPR101-Gs complex
| Entire | Name: Cryo-EM structure of the GPR101-Gs complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the GPR101-Gs complex
| Supramolecule | Name: Cryo-EM structure of the GPR101-Gs complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Soluble cytochrome b562,Probable G-protein coupled receptor 101
| Macromolecule | Name: Soluble cytochrome b562,Probable G-protein coupled receptor 101 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.588859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHENLYFQ GADLEDNWET LNDNLKVIEK ADNAAQVKDA LTKMRAAALD AQKATPPKL EDKSPDSPEM KDFRHGFDIL VGQIDDALKL ANEGKVKEAQ AAAEQLKTTR NAYIQKYLMT STCTNSTRES N SSHTCMPL ...String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHENLYFQ GADLEDNWET LNDNLKVIEK ADNAAQVKDA LTKMRAAALD AQKATPPKL EDKSPDSPEM KDFRHGFDIL VGQIDDALKL ANEGKVKEAQ AAAEQLKTTR NAYIQKYLMT STCTNSTRES N SSHTCMPL SKMPISLAHG IIRSTVLVIF LAASFVGNIV LALVLQRKPQ LLQVTNRFIF NLLVTDLLQI SLVAPWVVAT SV PLFWPLN SHFCTALVSL THLFAFASVN TIVVVSVDRY LSIIHPLSYP SKMTQRRGYL LLYGTWIVAI LQSTPPLYGW GQA AFDERN ALCSMIWGAS PSYTILSVVS FIVIPLIVMI ACYSVVFCAA RRQHALLYNV KRHSLEVRVK DCVENEDEEG AEKK EEFQD ESEFRRQHEG EVKAKEGRME AKDGSLKAKE GSTGTSESSV EARGSEEVRE SSTVASDGSM EGKEGSTKVE ENSMK ADKG RTEVNQCSID LGEDDMEFGE DDINFSEDDV EAVNIPESLP PSRRNSNSNP PLPRCYQCKA AKVIFIIIFS YVLSLG PYC FLAVLAVWVD VETQVPQWVI TIIIWLFFLQ CCIHPYVYGY MHKTIKKEIQ DMLKKFFCKE KPPKEDSHPD LPGTEGG TE GKIVPSYDSA TFP UniProtKB: Soluble cytochrome b562, Probable G-protein coupled receptor 101 |
-Macromolecule #2: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short
| Macromolecule | Name: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.69941 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQF RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE ...String: MGCLGNSKTE DQRNEEKAQR EANKKIEKQL QKDKQVYRAT HRLLLLGAGE SGKNTIVKQF RILHVNGFNG EGGEEDPQAA RSNSDGEKA TKVQDIKNNL KEAIETIVAA MSNLVPPVEL ANPENQFRVD YILSVMNVPD FDFPPEFYEH AKALWEDEGV R ACYERSNE YQLIDCAQYF LDKIDVIKQA DYVPSDQDLL RCRVLTSGIF ETKFQVDKVN FHMFDVGAQR DERRKWIQCF ND VTAIIFV VASSSYNMVI REDNQTNRLQ AALKLFDSIW NNKWLRDTSV ILFLNKQDLL AEKVLAGKSK IEDYFPEFAR YTT PEDATP EPGEDPRVTR AKYFIRDEFL RISTASGDGR HYCYPHFTCA VDTENIRRVF NDCRDIIQRM HLRQYELL UniProtKB: Guanine nucleotide-binding protein G(s) subunit alpha isoforms short |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.285734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW ...String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NAICFFPNGN AF ATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGHDNRVSCL GVT DDGMAV ATGSWDSFLK IWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 6.375332 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NTASIAQARK LVEQLKMEAN IDRIKVSKAA ADLMAYCEAH AKEDPLLTPV PASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Nb35
| Macromolecule | Name: Nb35 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.885439 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQESGGG LVQPGGSLRL SCAASGFTFS NYKMNWVRQA PGKGLEWVSD ISQSGASISY TGSVKGRFTI SRDNAKNTLY LQMNSLKPE DTAVYYCARC PAPFTRDCFD VTSTTYAYRG QGTQVTVSS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)