+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3717 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of differently sized plant protein | |||||||||
 Map data Map data | IM30 in C15 symmetrical ring assembly | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Saur M / Hennig R / Young P / Rusitzka K / Hellmann N / Heidrich J / Morgner N / Markl J / Schneider D | |||||||||
 Citation Citation |  Journal: Structure / Year: 2017 Journal: Structure / Year: 2017Title: A Janus-Faced IM30 Ring Involved in Thylakoid Membrane Fusion Is Assembled from IM30 Tetramers. Authors: Michael Saur / Raoul Hennig / Phoebe Young / Kristiane Rusitzka / Nadja Hellmann / Jennifer Heidrich / Nina Morgner / Jürgen Markl / Dirk Schneider /  Abstract: Biogenesis and dynamics of thylakoid membranes likely involves membrane fusion events. Membrane attachment of the inner membrane-associated protein of 30 kDa (IM30) affects the structure of the ...Biogenesis and dynamics of thylakoid membranes likely involves membrane fusion events. Membrane attachment of the inner membrane-associated protein of 30 kDa (IM30) affects the structure of the lipid bilayer, finally resulting in membrane fusion. Yet, how IM30 triggers membrane fusion is largely unclear. IM30 monomers pre-assemble into stable tetrameric building blocks, which further align to form oligomeric ring structures, and differently sized IM30 rings bind to membranes. Based on a 3D reconstruction of IM30 rings, we locate the IM30 loop 2 region at the bottom of the ring and show intact membrane binding but missing fusogenic activity of loop 2 mutants. However, helix 7, which has recently been shown to mediate membrane binding, was located at the oppossite, top side of IM30 rings. We propose that a two-sided IM30 ring complex connects two opposing membranes, finally resulting in membrane fusion. Thus, IM30-mediated membrane fusion requires a Janus-faced IM30 ring. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3717.map.gz emd_3717.map.gz | 907.3 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3717-v30.xml emd-3717-v30.xml emd-3717.xml emd-3717.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3717.png emd_3717.png | 141.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3717 http://ftp.pdbj.org/pub/emdb/structures/EMD-3717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3717 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3717 | HTTPS FTP |
-Validation report
| Summary document |  emd_3717_validation.pdf.gz emd_3717_validation.pdf.gz | 200.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3717_full_validation.pdf.gz emd_3717_full_validation.pdf.gz | 199.7 KB | Display | |
| Data in XML |  emd_3717_validation.xml.gz emd_3717_validation.xml.gz | 5.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3717 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3717 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3717 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3717 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3717.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3717.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | IM30 in C15 symmetrical ring assembly | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
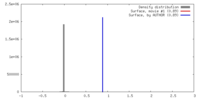
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Structure of a C15-symmetrical IM30 ring assembly
| Entire | Name: Structure of a C15-symmetrical IM30 ring assembly |
|---|---|
| Components |
|
-Supramolecule #1: Structure of a C15-symmetrical IM30 ring assembly
| Supramolecule | Name: Structure of a C15-symmetrical IM30 ring assembly / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 1.8 MDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 4503 |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera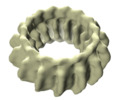






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)