+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rpd3S histone deacetylase complex | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | Rpd3S / HDAC / Hho1 / cryptic transcription / TRANSCRIPTION | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnucleosome disassembly/reassembly complex / negative regulation of antisense RNA transcription / Snt2C complex / negative regulation of reciprocal meiotic recombination / negative regulation of silent mating-type cassette heterochromatin formation / Rpd3L complex / protein localization to nucleolar rDNA repeats / Rpd3L-Expanded complex / negative regulation of rDNA heterochromatin formation / Rpd3S complex ...nucleosome disassembly/reassembly complex / negative regulation of antisense RNA transcription / Snt2C complex / negative regulation of reciprocal meiotic recombination / negative regulation of silent mating-type cassette heterochromatin formation / Rpd3L complex / protein localization to nucleolar rDNA repeats / Rpd3L-Expanded complex / negative regulation of rDNA heterochromatin formation / Rpd3S complex / rDNA chromatin condensation / regulation of RNA stability / nucleophagy / HDACs deacetylate histones / DNA replication-dependent chromatin assembly / histone deacetylase / SUMOylation of chromatin organization proteins / regulation of DNA-templated DNA replication initiation / nucleosome disassembly / cellular response to nitrogen starvation / negative regulation of transcription by RNA polymerase I / histone deacetylase activity / NuA4 histone acetyltransferase complex / Sin3-type complex / Estrogen-dependent gene expression / positive regulation of macroautophagy / histone deacetylase complex / histone acetyltransferase complex / methylated histone binding / nuclear periphery / meiotic cell cycle / positive regulation of transcription elongation by RNA polymerase II / transcription elongation by RNA polymerase II / heterochromatin formation / double-strand break repair via nonhomologous end joining / G1/S transition of mitotic cell cycle / transcription corepressor activity / G2/M transition of mitotic cell cycle / nucleosome assembly / cellular response to heat / response to oxidative stress / transcription coactivator activity / cell division / DNA repair / negative regulation of DNA-templated transcription / DNA-templated transcription / regulation of DNA-templated transcription / chromatin / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / identical protein binding / nucleus / metal ion binding / cytoplasm Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | |||||||||||||||||||||||||||
 Authors Authors | Dong S / Li H / Wang M / Rasheed N / Zou B / Gao X / Guan J / Li W / Zhang J / Wang C ...Dong S / Li H / Wang M / Rasheed N / Zou B / Gao X / Guan J / Li W / Zhang J / Wang C / Zhou N / Shi X / Li M / Zhou M / Huang J / Zhang Y / Wong KH / Zhang X / Chao WCH / Jun H | |||||||||||||||||||||||||||
| Funding support |  China, 8 items China, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex. Authors: Shuqi Dong / Huadong Li / Meilin Wang / Nadia Rasheed / Binqian Zou / Xijie Gao / Jiali Guan / Weijie Li / Jiale Zhang / Chi Wang / Ningkun Zhou / Xue Shi / Mei Li / Min Zhou / Junfeng Huang ...Authors: Shuqi Dong / Huadong Li / Meilin Wang / Nadia Rasheed / Binqian Zou / Xijie Gao / Jiali Guan / Weijie Li / Jiale Zhang / Chi Wang / Ningkun Zhou / Xue Shi / Mei Li / Min Zhou / Junfeng Huang / He Li / Ying Zhang / Koon Ho Wong / Xiaofei Zhang / William Chong Hang Chao / Jun He /  Abstract: In Saccharomyces cerevisiae, cryptic transcription at the coding region is prevented by the activity of Sin3 histone deacetylase (HDAC) complex Rpd3S, which is carried by the transcribing RNA ...In Saccharomyces cerevisiae, cryptic transcription at the coding region is prevented by the activity of Sin3 histone deacetylase (HDAC) complex Rpd3S, which is carried by the transcribing RNA polymerase II (RNAPII) to deacetylate and stabilize chromatin. Despite its fundamental importance, the mechanisms by which Rpd3S deacetylates nucleosomes and regulates chromatin dynamics remain elusive. Here, we determined several cryo-EM structures of Rpd3S in complex with nucleosome core particles (NCPs), including the H3/H4 deacetylation states, the alternative deacetylation state, the linker tightening state, and a state in which Rpd3S co-exists with the Hho1 linker histone on NCP. These structures suggest that Rpd3S utilizes a conserved Sin3 basic surface to navigate through the nucleosomal DNA, guided by its interactions with H3K36 methylation and the extra-nucleosomal DNA linkers, to target acetylated H3K9 and sample other histone tails. Furthermore, our structures illustrate that Rpd3S reconfigures the DNA linkers and acts in concert with Hho1 to engage the NCP, potentially unraveling how Rpd3S and Hho1 work in tandem for gene silencing. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37096.map.gz emd_37096.map.gz | 68 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37096-v30.xml emd-37096-v30.xml emd-37096.xml emd-37096.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37096.png emd_37096.png | 28.5 KB | ||
| Masks |  emd_37096_msk_1.map emd_37096_msk_1.map | 137.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37096.cif.gz emd-37096.cif.gz | 7.7 KB | ||
| Others |  emd_37096_half_map_1.map.gz emd_37096_half_map_1.map.gz emd_37096_half_map_2.map.gz emd_37096_half_map_2.map.gz | 127.4 MB 127.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37096 http://ftp.pdbj.org/pub/emdb/structures/EMD-37096 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37096 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37096 | HTTPS FTP |
-Validation report
| Summary document |  emd_37096_validation.pdf.gz emd_37096_validation.pdf.gz | 811.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37096_full_validation.pdf.gz emd_37096_full_validation.pdf.gz | 810.7 KB | Display | |
| Data in XML |  emd_37096_validation.xml.gz emd_37096_validation.xml.gz | 14.2 KB | Display | |
| Data in CIF |  emd_37096_validation.cif.gz emd_37096_validation.cif.gz | 16.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37096 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37096 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37096 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37096 | HTTPS FTP |
-Related structure data
| Related structure data |  8kc7MC  8kd2C  8kd3C  8kd4C  8kd5C  8kd6C  8kd7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37096.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37096.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
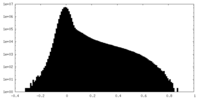
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37096_msk_1.map emd_37096_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
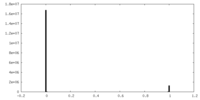
| Density Histograms |
-Half map: #2
| File | emd_37096_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
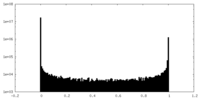
| Density Histograms |
-Half map: #1
| File | emd_37096_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
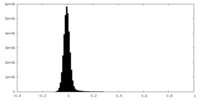
| Density Histograms |
- Sample components
Sample components
-Entire : Histone deacetylase complex Rpd3S in its apo form
| Entire | Name: Histone deacetylase complex Rpd3S in its apo form |
|---|---|
| Components |
|
-Supramolecule #1: Histone deacetylase complex Rpd3S in its apo form
| Supramolecule | Name: Histone deacetylase complex Rpd3S in its apo form / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 527.7 KDa |
-Macromolecule #1: Histone deacetylase RPD3
| Macromolecule | Name: Histone deacetylase RPD3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: histone deacetylase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.961957 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVYEATPFDP ITVKPSDKRR VAYFYDADVG NYAYGAGHPM KPHRIRMAHS LIMNYGLYKK MEIYRAKPAT KQEMCQFHTD EYIDFLSRV TPDNLEMFKR ESVKFNVGDD CPVFDGLYEY CSISGGGSME GAARLNRGKC DVAVNYAGGL HHAKKSEASG F CYLNDIVL ...String: MVYEATPFDP ITVKPSDKRR VAYFYDADVG NYAYGAGHPM KPHRIRMAHS LIMNYGLYKK MEIYRAKPAT KQEMCQFHTD EYIDFLSRV TPDNLEMFKR ESVKFNVGDD CPVFDGLYEY CSISGGGSME GAARLNRGKC DVAVNYAGGL HHAKKSEASG F CYLNDIVL GIIELLRYHP RVLYIDIDVH HGDGVEEAFY TTDRVMTCSF HKYGEFFPGT GELRDIGVGA GKNYAVNVPL RD GIDDATY RSVFEPVIKK IMEWYQPSAV VLQCGGDSLS GDRLGCFNLS MEGHANCVNY VKSFGIPMMV VGGGGYTMRN VAR TWCFET GLLNNVVLDK DLPYNEYYEY YGPDYKLSVR PSNMFNVNTP EYLDKVMTNI FANLENTKYA PSVQLNHTPR DAED LGDVE EDSAEAKDTK GGSQYARDLH VEHDNEFY UniProtKB: Histone deacetylase RPD3 |
-Macromolecule #2: Transcriptional regulatory protein SIN3
| Macromolecule | Name: Transcriptional regulatory protein SIN3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 157.586844 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHHHP QLAMWSHPQF EKGGGSGGGS GGGSWSHPQF EKENLYFQSD YRPLNVKDAL SYLEQVKFQF SSRPDIYNLF LDIMKDFKS QAIDTPGVIE RVSTLFRGYP ILIQGFNTFL PQGYRIECSS NPDDPIRVTT PMGTTTVNNN ISPSGRGTTD A QELGSFPE ...String: MHHHHHHHHP QLAMWSHPQF EKGGGSGGGS GGGSWSHPQF EKENLYFQSD YRPLNVKDAL SYLEQVKFQF SSRPDIYNLF LDIMKDFKS QAIDTPGVIE RVSTLFRGYP ILIQGFNTFL PQGYRIECSS NPDDPIRVTT PMGTTTVNNN ISPSGRGTTD A QELGSFPE SDGNGVQQPS NVPMVPSSVY QSEQNQDQQQ SLPLLATSSG LPSIQQPEMP AHRQIPQSQS LVPQEDAKKN VD VEFSQAI SYVNKIKTRF ADQPDIYKHF LEILQTYQRE QKPINEVYAQ VTHLFQNAPD LLEDFKKFLP DSSASANQQV QHA QQHAQQ QHEAQMHAQA QAQAQAQAQV EQQKQQQQFL YPASGYYGHP SNRGIPQQNL PPIGSFSPPT NGSTVHEAYQ DQQH MQPPH FMPLPSIVQH GPNMVHQGIA NENPPLSDLR TSLTEQYAPS SIQHQQQHPQ SISPIANTQY GDIPVRPEID LDPSI VPVV PEPTEPIENN ISLNEEVTFF EKAKRYIGNK HLYTEFLKIL NLYSQDILDL DDLVEKVDFY LGSNKELFTW FKNFVG YQE KTKCIENIVH EKHRLDLDLC EAFGPSYKRL PKSDTFMPCS GRDDMCWEVL NDEWVGHPVW ASEDSGFIAH RKNQYEE TL FKIEEERHEY DFYIESNLRT IQCLETIVNK IENMTENEKA NFKLPPGLGH TSMTIYKKVI RKVYDKERGF EIIDALHE H PAVTAPVVLK RLKQKDEEWR RAQREWNKVW RELEQKVFFK SLDHLGLTFK QADKKLLTTK QLISEISSIK VDQTNKKIH WLTPKPKSQL DFDFPDKNIF YDILCLADTF ITHTTAYSNP DKERLKDLLK YFISLFFSIS FEKIEESLYS HKQNVSESSG SDDGSSIAS RKRPYQQEMS LLDILHRSRY QKLKRSNDED GKVPQLSEPP EEEPNTIEEE ELIDEEAKNP WLTGNLVEEA N SQGIIQNR SIFNLFANTN IYIFFRHWTT IYERLLEIKQ MNERVTKEIN TRSTVTFAKD LDLLSSQLSE MGLDFVGEDA YK QVLRLSR RLINGDLEHQ WFEESLRQAY NNKAFKLYTI DKVTQSLVKH AHTLMTDAKT AEIMALFVKD RNASTTSAKD QII YRLQVR SHMSNTENMF RIEFDKRTLH VSIQYIALDD LTLKEPKADE DKWKYYVTSY ALPHPTEGIP HEKLKIPFLE RLIE FGQDI DGTEVDEEFS PEGISVSTLK IKIQPITYQL HIENGSYDVF TRKATNKYPT IANDNTQKGM VSQKKELISK FLDCA VGLR NNLDEAQKLS MQKKWENLKD SIAKTSAGNQ GIESETEKGK ITKQEQSDNL DSSTASVLPA SITTVPQDDN IETTGN TES SDKGAKIQ UniProtKB: Transcriptional regulatory protein SIN3 |
-Macromolecule #3: Chromatin modification-related protein EAF3
| Macromolecule | Name: Chromatin modification-related protein EAF3 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.266406 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVDLEQEFAL GGRCLAFHGP LMYEAKILKI WDPSSKMYTS IPNDKPGGSS QATKEIKPQK LGEDESIPEE IINGKCFFIH YQGWKSSWD EWVGYDRIRA YNEENIAMKK RLANEAKEAK KSLLEQQKKK KLSTSLGGPS NGGKRKGDSR SNASISKSTS Q SFLTSSVS ...String: MVDLEQEFAL GGRCLAFHGP LMYEAKILKI WDPSSKMYTS IPNDKPGGSS QATKEIKPQK LGEDESIPEE IINGKCFFIH YQGWKSSWD EWVGYDRIRA YNEENIAMKK RLANEAKEAK KSLLEQQKKK KLSTSLGGPS NGGKRKGDSR SNASISKSTS Q SFLTSSVS GRKSGRSSAN SLHPGSSLRS SSDQNGNDDR RRSSSLSPNM LHHIAGYPTP KISLQIPIKL KSVLVDDWEY VT KDKKICR LPADVTVEMV LNKYEHEVSQ ELESPGSQSQ LSEYCAGLKL YFDKCLGNML LYRLERLQYD ELLKKSSKDQ KPL VPIRIY GAIHLLRLIS VLPELISSTT MDLQSCQLLI KQTEDFLVWL LMHVDEYFND KDPNRSDDAL YVNTSSQYEG VALG M UniProtKB: Chromatin modification-related protein EAF3 |
-Macromolecule #4: Transcriptional regulatory protein RCO1
| Macromolecule | Name: Transcriptional regulatory protein RCO1 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 84.469328 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDTSKKDTTR SPSHSNSSSP SSSSLSSSSS KEKKRPKRLS SQNVNYDLKR RKIITSEGIE RSFKNEHSNL AVEDNIPEEE PKELLEKDS KGNIIKLNEP STISEDSKVS VTGLPLNKGP SEKIKRESLW NYRKNLGGQS NNSEMTLVPS KRFTQVPKNF Q DLNRNDLK ...String: MDTSKKDTTR SPSHSNSSSP SSSSLSSSSS KEKKRPKRLS SQNVNYDLKR RKIITSEGIE RSFKNEHSNL AVEDNIPEEE PKELLEKDS KGNIIKLNEP STISEDSKVS VTGLPLNKGP SEKIKRESLW NYRKNLGGQS NNSEMTLVPS KRFTQVPKNF Q DLNRNDLK TFLTENMTEE SNIRSTIGWN GDIINRTRDR EPESDRDNKK LSNIRTKIIL STNATYDSKS KLFGQNSIKS TS NASEKIF RDKNNSTIDF ENEDFCSACN QSGSFLCCDT CPKSFHFLCL DPPIDPNNLP KGDWHCNECK FKIFINNSMA TLK KIESNF IKQNNNVKIF AKLLFNIDSH NPKQFQLPNY IKETFPAVKT GSRGQYSDEN DKIPLTDRQL FNTSYGQSIT KLDS YNPDT HIDSNSGKFL ICYKCNQTRL GSWSHPENSR LIMTCDYCQT PWHLDCVPRA SFKNLGSKWK CPLHSPTKVY KKIHH CQED NSVNYKVWKK QRLINKKNQL YYEPLQKIGY QNNGNIQIIP TTSHTDYDFN QDFKITQIDE NSIKYDFFDK IYKSKM VQK RKLFQFQESL IDKLVSNGSQ NGNSEDNMVK DIASLIYFQV SNNDKSSNNK SASKSNNLRK LWDLKELTNV VVPNELD SI QFNDFSSDEI KHLLYLKKII ESKPKEELLK FLNIENPENQ SEMHHHHHHH HPQLAMWSHP QFEKGGGSGG GSGGGSWS H PQFEKENLYF QS UniProtKB: Transcriptional regulatory protein RCO1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.46 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 287163 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)