[English] 日本語
 Yorodumi
Yorodumi- EMDB-37125: Rpd3S in complex with nucleosome with H3K36MLA modification and 1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
 Keywords Keywords | Rpd3S / HDAC / Hho1 / cryptic transcription / TRANSCRIPTION | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationTINTIN complex / negative regulation of antisense RNA transcription / Snt2C complex / negative regulation of silent mating-type cassette heterochromatin formation / negative regulation of reciprocal meiotic recombination / Rpd3L complex / protein localization to nucleolar rDNA repeats / negative regulation of rDNA heterochromatin formation / Rpd3L-Expanded complex / Rpd3S complex ...TINTIN complex / negative regulation of antisense RNA transcription / Snt2C complex / negative regulation of silent mating-type cassette heterochromatin formation / negative regulation of reciprocal meiotic recombination / Rpd3L complex / protein localization to nucleolar rDNA repeats / negative regulation of rDNA heterochromatin formation / Rpd3L-Expanded complex / Rpd3S complex / rDNA chromatin condensation / nucleophagy / regulation of RNA stability / HDACs deacetylate histones / histone deacetylase activity, hydrolytic mechanism / DNA replication-dependent chromatin assembly / histone deacetylase / nucleosome disassembly / cellular response to nitrogen starvation / SUMOylation of chromatin organization proteins / regulation of DNA-templated DNA replication initiation / histone deacetylase activity / negative regulation of transcription by RNA polymerase I / histone deacetylase complex / : / Sin3-type complex / NuA4 histone acetyltransferase complex / histone reader activity / Estrogen-dependent gene expression / histone acetyltransferase complex / positive regulation of macroautophagy / nuclear periphery / meiotic cell cycle / transcription elongation by RNA polymerase II / positive regulation of transcription elongation by RNA polymerase II / G1/S transition of mitotic cell cycle / double-strand break repair via nonhomologous end joining / G2/M transition of mitotic cell cycle / structural constituent of chromatin / transcription corepressor activity / heterochromatin formation / nucleosome / nucleosome assembly / cellular response to heat / response to oxidative stress / transcription coactivator activity / protein heterodimerization activity / cell division / DNA repair / negative regulation of DNA-templated transcription / DNA-templated transcription / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / DNA binding / zinc ion binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||||||||
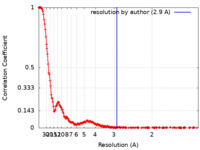
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||||||||||||||||||||
 Authors Authors | Dong S / Li H / Meilin W / Rasheed N / Zou B / Gao X / Guan J / Li W / Zhang J / Wang C ...Dong S / Li H / Meilin W / Rasheed N / Zou B / Gao X / Guan J / Li W / Zhang J / Wang C / Zhou N / Shi X / Li M / Zhou M / Huang J / Zhang Y / Wong KH / Zhang X / Chao WCH / He J | |||||||||||||||||||||||||||
| Funding support |  China, 8 items China, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex. Authors: Shuqi Dong / Huadong Li / Meilin Wang / Nadia Rasheed / Binqian Zou / Xijie Gao / Jiali Guan / Weijie Li / Jiale Zhang / Chi Wang / Ningkun Zhou / Xue Shi / Mei Li / Min Zhou / Junfeng Huang ...Authors: Shuqi Dong / Huadong Li / Meilin Wang / Nadia Rasheed / Binqian Zou / Xijie Gao / Jiali Guan / Weijie Li / Jiale Zhang / Chi Wang / Ningkun Zhou / Xue Shi / Mei Li / Min Zhou / Junfeng Huang / He Li / Ying Zhang / Koon Ho Wong / Xiaofei Zhang / William Chong Hang Chao / Jun He /  Abstract: In Saccharomyces cerevisiae, cryptic transcription at the coding region is prevented by the activity of Sin3 histone deacetylase (HDAC) complex Rpd3S, which is carried by the transcribing RNA ...In Saccharomyces cerevisiae, cryptic transcription at the coding region is prevented by the activity of Sin3 histone deacetylase (HDAC) complex Rpd3S, which is carried by the transcribing RNA polymerase II (RNAPII) to deacetylate and stabilize chromatin. Despite its fundamental importance, the mechanisms by which Rpd3S deacetylates nucleosomes and regulates chromatin dynamics remain elusive. Here, we determined several cryo-EM structures of Rpd3S in complex with nucleosome core particles (NCPs), including the H3/H4 deacetylation states, the alternative deacetylation state, the linker tightening state, and a state in which Rpd3S co-exists with the Hho1 linker histone on NCP. These structures suggest that Rpd3S utilizes a conserved Sin3 basic surface to navigate through the nucleosomal DNA, guided by its interactions with H3K36 methylation and the extra-nucleosomal DNA linkers, to target acetylated H3K9 and sample other histone tails. Furthermore, our structures illustrate that Rpd3S reconfigures the DNA linkers and acts in concert with Hho1 to engage the NCP, potentially unraveling how Rpd3S and Hho1 work in tandem for gene silencing. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37125.map.gz emd_37125.map.gz | 294.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37125-v30.xml emd-37125-v30.xml emd-37125.xml emd-37125.xml | 35.7 KB 35.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37125_fsc.xml emd_37125_fsc.xml | 24.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_37125.png emd_37125.png | 26.7 KB | ||
| Masks |  emd_37125_msk_1.map emd_37125_msk_1.map | 600.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37125.cif.gz emd-37125.cif.gz | 9.8 KB | ||
| Others |  emd_37125_half_map_1.map.gz emd_37125_half_map_1.map.gz emd_37125_half_map_2.map.gz emd_37125_half_map_2.map.gz | 553.5 MB 553.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37125 http://ftp.pdbj.org/pub/emdb/structures/EMD-37125 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37125 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37125 | HTTPS FTP |
-Related structure data
| Related structure data |  8kd5MC  8kc7C  8kd2C  8kd3C  8kd4C  8kd6C  8kd7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37125.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37125.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.71 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37125_msk_1.map emd_37125_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37125_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37125_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : H3/H4 deacetylation state of Rpd3S-NCP (187bp/MLA) class2
+Supramolecule #1: H3/H4 deacetylation state of Rpd3S-NCP (187bp/MLA) class2
+Supramolecule #2: Rpd3-Sin3-Eaf3-Rco1
+Supramolecule #3: Histone
+Supramolecule #4: 187bp DNA
+Macromolecule #1: Chromatin modification-related protein EAF3
+Macromolecule #2: Histone H3
+Macromolecule #3: Histone H4
+Macromolecule #4: Histone H2A
+Macromolecule #5: Histone H2B 1.1
+Macromolecule #8: Histone deacetylase RPD3
+Macromolecule #9: Transcriptional regulatory protein SIN3
+Macromolecule #10: Transcriptional regulatory protein RCO1
+Macromolecule #6: 187bp DNA
+Macromolecule #7: 187bp DNA
+Macromolecule #11: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)