+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3693 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Arabis Mosaic Virus capsid protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Arabis mosaic virus (isolate Syrah) Arabis mosaic virus (isolate Syrah) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Lecorre F | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Arabis Mosaic Virus capsid protein Authors: Lecorre F / Trapani S / Laikeehim J / Bron P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3693.map.gz emd_3693.map.gz | 21 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3693-v30.xml emd-3693-v30.xml emd-3693.xml emd-3693.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3693_fsc.xml emd_3693_fsc.xml | 17.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_3693.png emd_3693.png | 335.1 KB | ||
| Others |  emd_3693_half_map_1.map.gz emd_3693_half_map_1.map.gz emd_3693_half_map_2.map.gz emd_3693_half_map_2.map.gz | 283.2 MB 283.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3693 http://ftp.pdbj.org/pub/emdb/structures/EMD-3693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3693 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3693 | HTTPS FTP |
-Related structure data
| Related structure data |  5nsi M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3693.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3693.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
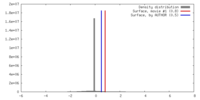
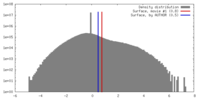
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_3693_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
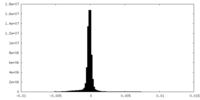
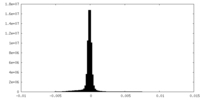
| Density Histograms |
-Half map: #1
| File | emd_3693_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
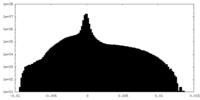
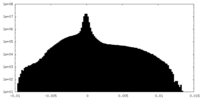
| Density Histograms |
- Sample components
Sample components
-Entire : Arabis mosaic virus (isolate Syrah)
| Entire | Name:  Arabis mosaic virus (isolate Syrah) Arabis mosaic virus (isolate Syrah) |
|---|---|
| Components |
|
-Supramolecule #1: Arabis mosaic virus (isolate Syrah)
| Supramolecule | Name: Arabis mosaic virus (isolate Syrah) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 283676 / Sci species name: Arabis mosaic virus (isolate Syrah) / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Rubus odoratus (flowering raspberry) Rubus odoratus (flowering raspberry) |
| Host (natural) | Organism:  Rheum rhabarbarum (garden rhubarb) Rheum rhabarbarum (garden rhubarb) |
| Host system | Organism:  Chenopodium quinoa (quinoa) Chenopodium quinoa (quinoa) |
-Macromolecule #1: RNA2 polyprotein
| Macromolecule | Name: RNA2 polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Arabis mosaic virus (isolate Syrah) / Strain: isolate Syrah Arabis mosaic virus (isolate Syrah) / Strain: isolate Syrah |
| Molecular weight | Theoretical: 55.765406 KDa |
| Recombinant expression | Organism:  Chenopodium quinoa (quinoa) Chenopodium quinoa (quinoa) |
| Sequence | String: GLAGRGTVPV PKDCQAGKYL KTLDLRDMVS GFSGIQYEKW ITAGLVMPDF KVVIRYPANA FTGITWVMSF DAYNRITSSI STTASPAYT LSVPHWLLHH KNGTTSCDID YGELCGHAMW FSATTFESPK LHFTCLTGNN KELAADWEFV VELYAEFEAA K SFLGKPNF ...String: GLAGRGTVPV PKDCQAGKYL KTLDLRDMVS GFSGIQYEKW ITAGLVMPDF KVVIRYPANA FTGITWVMSF DAYNRITSSI STTASPAYT LSVPHWLLHH KNGTTSCDID YGELCGHAMW FSATTFESPK LHFTCLTGNN KELAADWEFV VELYAEFEAA K SFLGKPNF IYSADAFNGS LKFLTIPPLE YDLSATSAYK SVSLLLGQTL VDGTHKVYNF NNTLLSYYLG IGGIVKGKVH VC SPCTYGI VLRVVSEWNG VTNNWNQLFK YPGCYIEEDG SFAIEIRSPY HRTPLRLIDA QSASSFTSTL NFYAISGPIA PSG ETAKMP VVVQIEEIAL PDLSVPSFPN DYFLWVDFSS FTVDVEEYVI GSRFFDISST TSTVALGDNP FSHMIACHGL HHGI LDLKL MWDLEGEFGK SSGGVTITKL CGDKATGMDG ASRVCALQNM GCETELYIGN YAGANPNTAL SLYSRWLAIK LDKAK SMKM LRILCKPRGN FEFYGRTCFK V |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average exposure time: 2.0 sec. / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model | 
PDB-5nsi: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)