[English] 日本語
 Yorodumi
Yorodumi- EMDB-9706: Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbe... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9706 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP | |||||||||
 Map data Map data | Cryo-EM map of MrNV semi-empty VLP | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | virion component / Viral coat protein subunit / Capsid protein alpha Function and homology information Function and homology information | |||||||||
| Biological species |  Macrobrachium rosenbergii nodavirus / Macrobrachium rosenbergii nodavirus /  Viruses Viruses | |||||||||
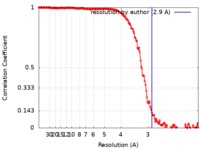
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Wang CH / Lin HH / Wu YY / Chang WH | |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP Authors: Wang CH / Lin HH / Wu YY / Chang WH | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9706.map.gz emd_9706.map.gz | 326.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9706-v30.xml emd-9706-v30.xml emd-9706.xml emd-9706.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9706_fsc.xml emd_9706_fsc.xml | 15.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_9706.png emd_9706.png | 357.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9706 http://ftp.pdbj.org/pub/emdb/structures/EMD-9706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9706 | HTTPS FTP |
-Related structure data
| Related structure data |  6jjcMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9706.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9706.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of MrNV semi-empty VLP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.026 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
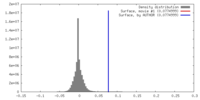
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Viruses
| Entire | Name:  Viruses Viruses |
|---|---|
| Components |
|
-Supramolecule #1: Viruses
| Supramolecule | Name: Viruses / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10239 / Sci species name: Viruses / Sci species strain: Taiwan / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Host system | Organism:  Macrobrachium rosenbergii nodavirus Macrobrachium rosenbergii nodavirus |
| Molecular weight | Theoretical: 7.452 MDa |
| Virus shell | Shell ID: 1 / Name: capsid protein [Macrobrachium rosenbergii nodavirus] / Diameter: 400.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: Capsid protein of giant freshwater prawn Macrobrachium rosenbergi...
| Macromolecule | Name: Capsid protein of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Macrobrachium rosenbergii nodavirus Macrobrachium rosenbergii nodavirus |
| Sequence | String: MARGKQNSNQ IQNNSNANGK RRKRNRRNRN PQTVPNFNPI VAKPTVAPLQ TNIRSARSDV NAITVLNGSD FLTTVKVRGS NNLIDSKSRI LVKQPISASS FLGTRISGLS QFWERYRWHK AAVRYVPAVP NTLACQLIGY IDTDPLDDPN VILDVDQLLR QATSQVGARQ ...String: MARGKQNSNQ IQNNSNANGK RRKRNRRNRN PQTVPNFNPI VAKPTVAPLQ TNIRSARSDV NAITVLNGSD FLTTVKVRGS NNLIDSKSRI LVKQPISASS FLGTRISGLS QFWERYRWHK AAVRYVPAVP NTLACQLIGY IDTDPLDDPN VILDVDQLLR QATSQVGARQ WNFSDTTTIP LIVRRDDQLY YTGQDKENVR FSQQGVFYLL QVTTLLNISG EAITNDLISG SLYLDWVCGF SMPQINPSPV EVSQLTYNAD TIGNWVPPTE LKQTYTQDIT GLKPNSKFII IPYMDRVSSE VLQKCTITCN EVDAVGSISY SDTSAIKCDG YILFQANSIG EATFTLVTDY QGAVDPKPYQ YRIIRAIVGN N |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 270 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 93.0 K / Max: 103.0 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 37.0 e/Å2 Details: Images were collected in movie-mode with 30 frames within 3 second |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus min: 0.45 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.37 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)