[English] 日本語
 Yorodumi
Yorodumi- EMDB-36885: Cryo-EM structure of 30S ribosome with cleaved AP-mRNA bound comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of 30S ribosome with cleaved AP-mRNA bound complex-II (Body 1) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | 30 Ribosome / AP-mRNA / RIBOSOME | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||
 Authors Authors | Ramachandran R / Afsar M / Shukla A | ||||||||||||
| Funding support |  India, 3 items India, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Bacterial Rps3 counters oxidative and UV stress by recognizing and processing AP-sites on mRNA via a novel mechanism. Authors: Mohammad Afsar / Ankita Shukla / Faiz Ali / Rahul Kumar Maurya / Suman Bharti / Nelam Kumar / Mohammad Sadik / Surabhi Chandra / Huma Rahil / Sanjay Kumar / Imran Ansari / Farheen Jahan / ...Authors: Mohammad Afsar / Ankita Shukla / Faiz Ali / Rahul Kumar Maurya / Suman Bharti / Nelam Kumar / Mohammad Sadik / Surabhi Chandra / Huma Rahil / Sanjay Kumar / Imran Ansari / Farheen Jahan / Saman Habib / Tanweer Hussain / Manju Yasoda Krishnan / Ravishankar Ramachandran /  Abstract: Lesions and stable secondary structures in mRNA severely impact the translation efficiency, causing ribosome stalling and collisions. Prokaryotic ribosomal proteins Rps3, Rps4 and Rps5, located in ...Lesions and stable secondary structures in mRNA severely impact the translation efficiency, causing ribosome stalling and collisions. Prokaryotic ribosomal proteins Rps3, Rps4 and Rps5, located in the mRNA entry tunnel, form the mRNA helicase center and unwind stable mRNA secondary structures during translation. However, the mechanism underlying the detection of lesions on translating mRNA is unclear. We used Cryo-EM, biochemical assays, and knockdown experiments to investigate the apurinic/apyrimidinic (AP) endoribonuclease activity of bacterial ribosomes on AP-site containing mRNA. Our biochemical assays show that Rps3, specifically the 130RR131 motif, is important for recognizing and performing the AP-endoribonuclease activity. Furthermore, structural analysis revealed cleaved mRNA product in the 30S ribosome entry tunnel. Additionally, knockdown studies in Mycobacterium tuberculosis confirmed the protective role of Rps3 against oxidative and UV stress. Overall, our results show that prokaryotic Rps3 recognizes and processes AP-sites on mRNA via a novel mechanism that is distinct from eukaryotes. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36885.map.gz emd_36885.map.gz | 45.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36885-v30.xml emd-36885-v30.xml emd-36885.xml emd-36885.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36885.png emd_36885.png | 79.3 KB | ||
| Masks |  emd_36885_msk_1.map emd_36885_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36885.cif.gz emd-36885.cif.gz | 4.2 KB | ||
| Others |  emd_36885_half_map_1.map.gz emd_36885_half_map_1.map.gz emd_36885_half_map_2.map.gz emd_36885_half_map_2.map.gz | 45.5 MB 45.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36885 http://ftp.pdbj.org/pub/emdb/structures/EMD-36885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36885 | HTTPS FTP |
-Validation report
| Summary document |  emd_36885_validation.pdf.gz emd_36885_validation.pdf.gz | 827.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36885_full_validation.pdf.gz emd_36885_full_validation.pdf.gz | 827 KB | Display | |
| Data in XML |  emd_36885_validation.xml.gz emd_36885_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_36885_validation.cif.gz emd_36885_validation.cif.gz | 14.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36885 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36885 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36885 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36885 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36885.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36885.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||
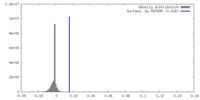
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36885_msk_1.map emd_36885_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
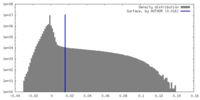
| Density Histograms |
-Half map: #1
| File | emd_36885_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36885_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 30S Ribosome+AP-mRNA
| Entire | Name: 30S Ribosome+AP-mRNA |
|---|---|
| Components |
|
-Supramolecule #1: 30S Ribosome+AP-mRNA
| Supramolecule | Name: 30S Ribosome+AP-mRNA / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 41.53 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 42320 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)