[English] 日本語
 Yorodumi
Yorodumi- EMDB-36871: In situ structure of RNA-dependent RNA polymerase in full BAV par... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ structure of RNA-dependent RNA polymerase in full BAV particles | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | reovirus / RdRp / complex / inner capsid protein / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral genome replication / RNA-directed RNA polymerase / nucleotide binding / RNA-directed RNA polymerase activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  Banna virus Banna virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Li Z / Cao S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes. Authors: Zhiqiang Li / Han Xia / Guibo Rao / Yan Fu / Tingting Chong / Kexing Tian / Zhiming Yuan / Sheng Cao /  Abstract: Banna virus (BAV) is the prototype Seadornavirus, a class of reoviruses for which there has been little structural study. Here, we report atomic cryo-EM structures of three states of BAV virions- ...Banna virus (BAV) is the prototype Seadornavirus, a class of reoviruses for which there has been little structural study. Here, we report atomic cryo-EM structures of three states of BAV virions-surrounded by 120 spikes (full virions), 60 spikes (partial virions), or no spikes (cores). BAV cores are double-layered particles similar to the cores of other non-turreted reoviruses, except for an additional protein component in the outer capsid shell, VP10. VP10 was identified to be a cementing protein that plays a pivotal role in the assembly of BAV virions by directly interacting with VP2 (inner capsid), VP8 (outer capsid), and VP4 (spike). Viral spikes (VP4/VP9 heterohexamers) are situated on top of VP10 molecules in full or partial virions. Asymmetrical electrostatic interactions between VP10 monomers and VP4 trimers are disrupted by high pH treatment, which is thus a simple way to produce BAV cores. Low pH treatment of BAV virions removes only the flexible receptor binding protein VP9 and triggers significant conformational changes in the membrane penetration protein VP4. BAV virions adopt distinct spatial organization of their surface proteins compared with other well-studied reoviruses, suggesting that BAV may have a unique mechanism of penetration of cellular endomembranes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36871.map.gz emd_36871.map.gz | 306.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36871-v30.xml emd-36871-v30.xml emd-36871.xml emd-36871.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36871_fsc.xml emd_36871_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36871.png emd_36871.png | 176.7 KB | ||
| Filedesc metadata |  emd-36871.cif.gz emd-36871.cif.gz | 6.9 KB | ||
| Others |  emd_36871_half_map_1.map.gz emd_36871_half_map_1.map.gz emd_36871_half_map_2.map.gz emd_36871_half_map_2.map.gz | 277.4 MB 277.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36871 http://ftp.pdbj.org/pub/emdb/structures/EMD-36871 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36871 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36871 | HTTPS FTP |
-Validation report
| Summary document |  emd_36871_validation.pdf.gz emd_36871_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36871_full_validation.pdf.gz emd_36871_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_36871_validation.xml.gz emd_36871_validation.xml.gz | 23.5 KB | Display | |
| Data in CIF |  emd_36871_validation.cif.gz emd_36871_validation.cif.gz | 31.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36871 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36871 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36871 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36871 | HTTPS FTP |
-Related structure data
| Related structure data |  8k43MC  8k42C  8k44C  8k49C  8k4aC  8w9pC  8w9qC  8w9rC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36871.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36871.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
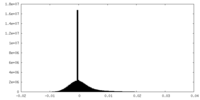
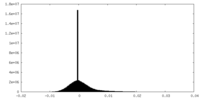
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36871_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
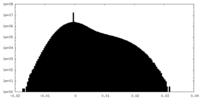
| Density Histograms |
-Half map: #1
| File | emd_36871_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
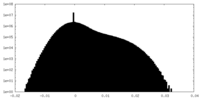
| Density Histograms |
- Sample components
Sample components
-Entire : Banna virus
| Entire | Name:  Banna virus Banna virus |
|---|---|
| Components |
|
-Supramolecule #1: Banna virus
| Supramolecule | Name: Banna virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 77763 / Sci species name: Banna virus / Sci species strain: YN15-126-01 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: RNA-directed RNA polymerase (Fragment)
| Macromolecule | Name: RNA-directed RNA polymerase (Fragment) / type: protein_or_peptide / ID: 1 / Details: OR004518 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Banna virus Banna virus |
| Molecular weight | Theoretical: 138.563344 KDa |
| Sequence | String: MDVQEQFEGY LREEVDLLNK FEDSHFKQLE IFYTKSQTDH VINKDKLQFN ALPFHTTLYK EINGKRVRLG TLLNWTKAER LDTIRHESM IKDERLRRLI DFDYGWIDYA VVLQRYLDEG NTIESANILQ FDGDFVNNIK HPTQKNNFLD IKVIKECETY I LMKDDNGI ...String: MDVQEQFEGY LREEVDLLNK FEDSHFKQLE IFYTKSQTDH VINKDKLQFN ALPFHTTLYK EINGKRVRLG TLLNWTKAER LDTIRHESM IKDERLRRLI DFDYGWIDYA VVLQRYLDEG NTIESANILQ FDGDFVNNIK HPTQKNNFLD IKVIKECETY I LMKDDNGI RDPDILRAWE LNSIPEVIEL DVDGETKKFN LRKEMIKRIQ DEAPVYFFCN PYRYAVANLD PNIPEVRLWL EY FIGSEDF FGFNGPNVIV SKSLLAAKRF EVVVNHLRKV AAFELDVESK EVMNEWIKYI MIDPVYRSYK NRGAFGNLNQ HVF ARTKSE GLSWSLIDIG TTNFELKPTK KVAGSYVNKF NLVDDVLVEE SLKDLRNEGL HKMADVTRRM IDADITPENV KKGK LNRLA LSYCGYTGSH SATAMVKQFN GTKDLDPNCD PMFVDIVKDN MRVYMQEGLQ KYPQGSRKSN RLDILFKGGT SSASS TNEH AVVNGRFRYR SELYRERDVN SSTVFKTATP GQYRVVKKIS AKLKSKNANI VTHPMNFINF KVDDLDIVVN AGSRLV RGT RAKRIITPNY GTIYAASLMT VLPAVRLLSS RASNMGALST QGRIGTTYHG ALPHDVMAPQ LAVTSSDDVS KICVAKD FG QFDTSQWGQI SKAHADGVRS MKAHYSMGHD TLVDLDLNDA SFADLLEVTA MSYERPLKYK MNGLVCESAG VKSGELTT Q TRNTTTNISH STVALDDYNN RAYRLNLPKL ELVTDNKVGD DSVEVLRVVD GSPLTPEIAK LYVNCMQDHA DKNHLEISA KRTIVGNNVA EHIKIWVFKG YLALDVFLDS VTSEKNSFSN LNYLEQVNIL YDMAMTLMIR YCSVQACMTQ FCNDMKLLNG IRAGNYTFI PTPKIICAYG TPEICLRAPE IRSFGRYLPI DEDEYSVLND LVASLSTNKP KMDFVAQMFE QNGNQVHGIW L DHFKRKND VNPDGGGIHI SEGLKRLMPE YCERHLNELV YKTLDDKVIR DYTSDIIITN ICKGKLSKAP KLAFFANFYL SL TGFNGVD SPYLTADEGV KNVHRVIGLS YRNTLSTSPT ANVDRILRNN PGSAPAYLTG NDILGVLSDY PYQNWRTVVE LLD ITEPSA TAIIEVATNQ MHAYLADKDL NTANLFDNTS RTYDISDRTY PKFVNITSNL SNSNRRGFQL EAMKHIIYMA RRGI ATLAN THPSKIGNTV YYDY UniProtKB: RNA-directed RNA polymerase |
-Macromolecule #2: VP2
| Macromolecule | Name: VP2 / type: protein_or_peptide / ID: 2 / Details: OR004519 / Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Banna virus Banna virus |
| Molecular weight | Theoretical: 108.031008 KDa |
| Sequence | String: MPRKKDQVTK NDDGNQTSDV QTQDFKTAVQ PDTNTAQLIK TYSNPKQRGD KGEIIYDGGL SSKLADVVDK TTEPHNADGA VKDGRIAPV KLDLEKQKLD KLKLFETSPF DPLTIKNNQD VVDKLYATQS SSIQEVVPTK TFATELQFGV TSEDMAKIYG A VAAVSKNV ...String: MPRKKDQVTK NDDGNQTSDV QTQDFKTAVQ PDTNTAQLIK TYSNPKQRGD KGEIIYDGGL SSKLADVVDK TTEPHNADGA VKDGRIAPV KLDLEKQKLD KLKLFETSPF DPLTIKNNQD VVDKLYATQS SSIQEVVPTK TFATELQFGV TSEDMAKIYG A VAAVSKNV NSSVTYEVKR GTHELIKVPT IPHNLVLIQS DNGKHALIKE DLGQWPVETG ISLVNQAGVF AVQLANKLGI DK PFVLDAG SNYFTDTSFI DTRKYCTDGL SPREIQKALN RQRAYYDRPE LTISENKTLL SQSIIYPDAD GNDVSIIFSG AMS HAIFTY AQSQWNKNII KLDDYIREIT LTVPKQYRPR RFKEIEHTHG YVYRELNQGS LLPLVDANLK ESSSYYFKKL MSSI SNVPV DARTLQSATA ALAADTGQAV NRAQHVSMLT NRLTTANAPT VRAITVLTCM FKQFRIGMTY ALDPNIMDVA AATCM LLFR PAQSISDEQY RYCLQTMAVF LTNTTYDIVN NDTIDVLKMK LRNQGWPFVE RYNAVEIDMS VEPLRSPGQV GRYYNP FNI DPLTKKHVED RLEEFINQVQ VGRFRNASGN AVGTTLAAFL RACRDKTSAN WRGYSVLVSR YRSLIPNELF ESLRNIS GE YNINPQDEHS FFFALAQINA DDEFIGAIDK ESAEYLDEYA TLARDISNSL TLVKAAFGPL ERTSGSIINH ANNLNKVI N HVFADKPLIS ETMLKILTID GTTGKDGYRN WLDKLVGHNY PVYVEPVVNI MNFISARFVA DSSYFGYTNE IMIMPNHIN VPVDDRFGFR DSPFCTSLPR TIMGNDVRRI SYNVFSMMED IDDVISEGFI LYDAYFNFSY DIMTTDGVTR LKEDILIVTD TGNDIKPIH FYIYFENRND KKLRYESKMN VSYRLYIKTP ACLLPLSDYM RAQHDYVSPS SSRVYIKDPA VVYTRS UniProtKB: Vp2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)