+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the bacteriophage lambda neck | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationvirus tail, tube / symbiont genome ejection through host cell envelope, long flexible tail mechanism / viral tail assembly / virus tail / virion assembly / virion component / host cell cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) | |||||||||||||||
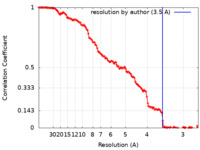
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||
 Authors Authors | Xiao H / Tan L / Cheng LP / Liu HR | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2023 Journal: PLoS Biol / Year: 2023Title: Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly. Authors: Hao Xiao / Le Tan / Zhixue Tan / Yewei Zhang / Wenyuan Chen / Xiaowu Li / Jingdong Song / Lingpeng Cheng / Hongrong Liu /  Abstract: Siphophages have a long, flexible, and noncontractile tail that connects to the capsid through a neck. The phage tail is essential for host cell recognition and virus-host cell interactions; ...Siphophages have a long, flexible, and noncontractile tail that connects to the capsid through a neck. The phage tail is essential for host cell recognition and virus-host cell interactions; moreover, it serves as a channel for genome delivery during infection. However, the in situ high-resolution structure of the neck-tail complex of siphophages remains unknown. Here, we present the structure of the siphophage lambda "wild type," the most widely used, laboratory-adapted fiberless mutant. The neck-tail complex comprises a channel formed by stacked 12-fold and hexameric rings and a 3-fold symmetrical tip. The interactions among DNA and a total of 246 tail protein molecules forming the tail and neck have been characterized. Structural comparisons of the tail tips, the most diversified region across the lambda and other long-tailed phages or tail-like machines, suggest that their tail tip contains conserved domains, which facilitate tail assembly, receptor binding, cell adsorption, and DNA retaining/releasing. These domains are distributed in different tail tip proteins in different phages or tail-like machines. The side tail fibers are not required for the phage particle to orient itself vertically to the surface of the host cell during attachment. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36846.map.gz emd_36846.map.gz | 164.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36846-v30.xml emd-36846-v30.xml emd-36846.xml emd-36846.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36846_fsc.xml emd_36846_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_36846.png emd_36846.png | 88.3 KB | ||
| Filedesc metadata |  emd-36846.cif.gz emd-36846.cif.gz | 5.3 KB | ||
| Others |  emd_36846_half_map_1.map.gz emd_36846_half_map_1.map.gz emd_36846_half_map_2.map.gz emd_36846_half_map_2.map.gz | 163.6 MB 163.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36846 http://ftp.pdbj.org/pub/emdb/structures/EMD-36846 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36846 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36846 | HTTPS FTP |
-Validation report
| Summary document |  emd_36846_validation.pdf.gz emd_36846_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36846_full_validation.pdf.gz emd_36846_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_36846_validation.xml.gz emd_36846_validation.xml.gz | 20.3 KB | Display | |
| Data in CIF |  emd_36846_validation.cif.gz emd_36846_validation.cif.gz | 26.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36846 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36846 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36846 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36846 | HTTPS FTP |
-Related structure data
| Related structure data |  8k37MC  8k35C  8k36C  8k38C  8k39C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36846.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36846.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36846_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36846_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage Lambda
| Entire | Name:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage Lambda
| Supramolecule | Name: Escherichia phage Lambda / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2681611 / Sci species name: Escherichia phage Lambda / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Tail tube protein
| Macromolecule | Name: Tail tube protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 25.831779 KDa |
| Sequence | String: MPVPNPTMPV KGAGTTLWVY KGSGDPYANP LSDVDWSRLA KVKDLTPGEL TAESYDDSYL DDEDADWTAT GQGQKSAGDT SFTLAWMPG EQGQQALLAW FNEGDTRAYK IRFPNGTVDV FRGWVSSIGK AVTAKEVITR TVKVTNVGRP SMAEDRSTVT A ATGMTVTP ...String: MPVPNPTMPV KGAGTTLWVY KGSGDPYANP LSDVDWSRLA KVKDLTPGEL TAESYDDSYL DDEDADWTAT GQGQKSAGDT SFTLAWMPG EQGQQALLAW FNEGDTRAYK IRFPNGTVDV FRGWVSSIGK AVTAKEVITR TVKVTNVGRP SMAEDRSTVT A ATGMTVTP ASTSVVKGQS TTLTVAFQPE GVTDKSFRAV SADKTKATVS VSGMTITVNG VAAGKVNIPV VSGNGEFAAV AE ITVTAS UniProtKB: Tail tube protein |
-Macromolecule #2: Tail tube terminator protein
| Macromolecule | Name: Tail tube terminator protein / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 14.659124 KDa |
| Sequence | String: MKHTELRAAV LDALEKHDTG ATFFDGRPAV FDEADFPAVA VYLTGAEYTG EELDSDTWQA ELHIEVFLPA QVPDSELDAW MESRIYPVM SDIPALSDLI TSMVASGYDY RRDDDAGLWS SADLTYVITY EM UniProtKB: Tail tube terminator protein |
-Macromolecule #3: Head-tail connector protein FII
| Macromolecule | Name: Head-tail connector protein FII / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 12.775037 KDa |
| Sequence | String: MADFDNLFDA AIARADETIR GYMGTSATIT SGEQSGAVIR GVFDDPENIS YAGQGVRVEG SSPSLFVRTD EVRQLRRGDT LTIGEENFW VDRVSPDDGG SCHLWLGRGV PPAVNRRR UniProtKB: Head-tail connector protein FII |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)