+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the bacteriophage lambda tail tip complex | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationvirus tail, tube / symbiont genome ejection through host cell envelope, long flexible tail mechanism / viral tail assembly / virus tail / 4 iron, 4 sulfur cluster binding / entry receptor-mediated virion attachment to host cell / host cell cytoplasm / receptor-mediated virion attachment to host cell / virion attachment to host cell / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.44 Å | |||||||||||||||
 Authors Authors | Xiao H / Tan L / Cheng LP / Liu HR | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2023 Journal: PLoS Biol / Year: 2023Title: Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly. Authors: Hao Xiao / Le Tan / Zhixue Tan / Yewei Zhang / Wenyuan Chen / Xiaowu Li / Jingdong Song / Lingpeng Cheng / Hongrong Liu /  Abstract: Siphophages have a long, flexible, and noncontractile tail that connects to the capsid through a neck. The phage tail is essential for host cell recognition and virus-host cell interactions; ...Siphophages have a long, flexible, and noncontractile tail that connects to the capsid through a neck. The phage tail is essential for host cell recognition and virus-host cell interactions; moreover, it serves as a channel for genome delivery during infection. However, the in situ high-resolution structure of the neck-tail complex of siphophages remains unknown. Here, we present the structure of the siphophage lambda "wild type," the most widely used, laboratory-adapted fiberless mutant. The neck-tail complex comprises a channel formed by stacked 12-fold and hexameric rings and a 3-fold symmetrical tip. The interactions among DNA and a total of 246 tail protein molecules forming the tail and neck have been characterized. Structural comparisons of the tail tips, the most diversified region across the lambda and other long-tailed phages or tail-like machines, suggest that their tail tip contains conserved domains, which facilitate tail assembly, receptor binding, cell adsorption, and DNA retaining/releasing. These domains are distributed in different tail tip proteins in different phages or tail-like machines. The side tail fibers are not required for the phage particle to orient itself vertically to the surface of the host cell during attachment. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36844.map.gz emd_36844.map.gz | 37.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36844-v30.xml emd-36844-v30.xml emd-36844.xml emd-36844.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36844.png emd_36844.png | 120.7 KB | ||
| Filedesc metadata |  emd-36844.cif.gz emd-36844.cif.gz | 7.7 KB | ||
| Others |  emd_36844_half_map_1.map.gz emd_36844_half_map_1.map.gz emd_36844_half_map_2.map.gz emd_36844_half_map_2.map.gz | 31.4 MB 31.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36844 http://ftp.pdbj.org/pub/emdb/structures/EMD-36844 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36844 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36844 | HTTPS FTP |
-Validation report
| Summary document |  emd_36844_validation.pdf.gz emd_36844_validation.pdf.gz | 880 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36844_full_validation.pdf.gz emd_36844_full_validation.pdf.gz | 879.5 KB | Display | |
| Data in XML |  emd_36844_validation.xml.gz emd_36844_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_36844_validation.cif.gz emd_36844_validation.cif.gz | 12.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36844 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36844 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36844 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36844 | HTTPS FTP |
-Related structure data
| Related structure data |  8k35MC  8k36C  8k37C  8k38C  8k39C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36844.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36844.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36844_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
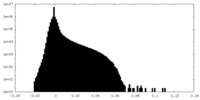
| Projections & Slices |
| ||||||||||||
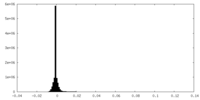
| Density Histograms |
-Half map: #1
| File | emd_36844_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
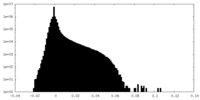
| Projections & Slices |
| ||||||||||||
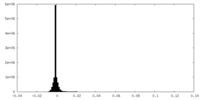
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage Lambda
| Entire | Name:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage Lambda
| Supramolecule | Name: Escherichia phage Lambda / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 / NCBI-ID: 2681611 / Sci species name: Escherichia phage Lambda / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Tip attachment protein J
| Macromolecule | Name: Tip attachment protein J / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 124.550625 KDa |
| Sequence | String: MGKGSSKGHT PREAKDNLKS TQLLSVIDAI SEGPIEGPVD GLKSVLLNST PVLDTEGNTN ISGVTVVFRA GEQEQTPPEG FESSGSETV LGTEVKYDTP ITRTITSANI DRLRFTFGVQ ALVETTSKGD RNPSEVRLLV QIQRNGGWVT EKDITIKGKT T SQYLASVV ...String: MGKGSSKGHT PREAKDNLKS TQLLSVIDAI SEGPIEGPVD GLKSVLLNST PVLDTEGNTN ISGVTVVFRA GEQEQTPPEG FESSGSETV LGTEVKYDTP ITRTITSANI DRLRFTFGVQ ALVETTSKGD RNPSEVRLLV QIQRNGGWVT EKDITIKGKT T SQYLASVV MGNLPPRPFN IRMRRMTPDS TTDQLQNKTL WSSYTEIIDV KQCYPNTALV GVQVDSEQFG SQQVSRNYHL RG RILQVPS NYNPQTRQYS GIWDGTFKPA YSNNMAWCLW DMLTHPRYGM GKRLGAADVD KWALYVIGQY CDQSVPDGFG GTE PRITCN AYLTTQRKAW DVLSDFCSAM RCMPVWNGQT LTFVQDRPSD KTWTYNRSNV VMPDDGAPFR YSFSALKDRH NAVE VNWID PNNGWETATE LVEDTQAIAR YGRNVTKMDA FGCTSRGQAH RAGLWLIKTE LLETQTVDFS VGAEGLRHVP GDVIE ICDD DYAGISTGGR VLAVNSQTRT LTLDREITLP SSGTALISLV DGSGNPVSVE VQSVTDGVKV KVSRVPDGVA EYSVWE LKL PTLRQRLFRC VSIRENDDGT YAITAVQHVP EKEAIVDNGA HFDGEQSGTV NGVTPPAVQH LTAEVTADSG EYQVLAR WD TPKVVKGVSF LLRLTVTADD GSERLVSTAR TTETTYRFTQ LALGNYRLTV RAVNAWGQQG DPASVSFRIA APAAPSRI E LTPGYFQITA TPHLAVYDPT VQFEFWFSEK QIADIRQVET STRYLGTALY WIAASINIKP GHDYYFYIRS VNTVGKSAF VEAVGRASDD AEGYLDFFKG KITESHLGKE LLEKVELTED NASRLEEFSK EWKDASDKWN AMWAVKIEQT KDGKHYVAGI GLSMEDTEE GKLSQFLVAA NRIAFIDPAN GNETPMFVAQ GNQIFMNDVF LKRLTAPTIT SGGNPPAFSL TPDGKLTAKN A DISGSVNA NSGTLSNVTI AENCTINGTL RAEKIVGDIV KAASAAFPRQ RESSVDWPSG TRTVTVTDDH PFDRQIVVLP LT FRGSKRT VSGRTTYSMC YLKVLMNGAV IYDGAANEAV QVFSRIVDMP AGRGNVILTF TLTSTRHSAD IPPYTFASDV QVM VIKKQA LGISVV UniProtKB: Tip attachment protein J |
-Macromolecule #2: Tail tip protein L
| Macromolecule | Name: Tail tip protein L / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 25.730578 KDa |
| Sequence | String: MQDIRQETLN ECTRAEQSAS VVLWEIDLTE VGGERYFFCN EQNEKGEPVT WQGRQYQPYP IQGSGFELNG KGTSTRPTLT VSNLYGMVT GMAEDMQSLV GGTVVRRKVY ARFLDAVNFV NGNSYADPEQ EVISRWRIEQ CSELSAVSAS FVLSTPTETD G AVFPGRIM ...String: MQDIRQETLN ECTRAEQSAS VVLWEIDLTE VGGERYFFCN EQNEKGEPVT WQGRQYQPYP IQGSGFELNG KGTSTRPTLT VSNLYGMVT GMAEDMQSLV GGTVVRRKVY ARFLDAVNFV NGNSYADPEQ EVISRWRIEQ CSELSAVSAS FVLSTPTETD G AVFPGRIM LANTCTWTYR GDECGYSGPA VADEYDQPTS DITKDKCSKC LSGCKFRNNV GNFGGFLSIN KLSQ UniProtKB: Tail tip protein L |
-Macromolecule #3: Tail tip protein M
| Macromolecule | Name: Tail tip protein M / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 12.547373 KDa |
| Sequence | String: MKTFRWKVKP GMDVASVPSV RKVRFGDGYS QRAPAGLNAN LKTYSVTLSV PREEATVLES FLEEHGGWKS FLWTPPYEWR QIKVTCAKW SSRVSMLRVE FSAEFEQVVN UniProtKB: Tail tip protein M |
-Macromolecule #4: Tail tip assembly protein I
| Macromolecule | Name: Tail tip assembly protein I / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 23.14659 KDa |
| Sequence | String: MAATHTLPLA SPGMARICLY GDLQRFGRRI DLRVKTGAEA IRALATQLPA FRQKLSDGWY QVRIAGRDVS TSGLTAQLHE TLPDGAVIH IVPRVAGAKS GGVFQIVLGA AAIAGSFFTA GATLAAWGAA IGAGGMTGIL FSLGASMVLG GVAQMLAPKA R TPRIQTTD ...String: MAATHTLPLA SPGMARICLY GDLQRFGRRI DLRVKTGAEA IRALATQLPA FRQKLSDGWY QVRIAGRDVS TSGLTAQLHE TLPDGAVIH IVPRVAGAKS GGVFQIVLGA AAIAGSFFTA GATLAAWGAA IGAGGMTGIL FSLGASMVLG GVAQMLAPKA R TPRIQTTD NGKQNTYFSS LDNMVAQGNV LPVLYGEMRV GSRVVSQEIS TADEGDGGQV VVIGR UniProtKB: Tail tip assembly protein I |
-Macromolecule #5: Tail tube protein
| Macromolecule | Name: Tail tube protein / type: protein_or_peptide / ID: 5 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 25.831779 KDa |
| Sequence | String: MPVPNPTMPV KGAGTTLWVY KGSGDPYANP LSDVDWSRLA KVKDLTPGEL TAESYDDSYL DDEDADWTAT GQGQKSAGDT SFTLAWMPG EQGQQALLAW FNEGDTRAYK IRFPNGTVDV FRGWVSSIGK AVTAKEVITR TVKVTNVGRP SMAEDRSTVT A ATGMTVTP ...String: MPVPNPTMPV KGAGTTLWVY KGSGDPYANP LSDVDWSRLA KVKDLTPGEL TAESYDDSYL DDEDADWTAT GQGQKSAGDT SFTLAWMPG EQGQQALLAW FNEGDTRAYK IRFPNGTVDV FRGWVSSIGK AVTAKEVITR TVKVTNVGRP SMAEDRSTVT A ATGMTVTP ASTSVVKGQS TTLTVAFQPE GVTDKSFRAV SADKTKATVS VSGMTITVNG VAAGKVNIPV VSGNGEFAAV AE ITVTAS UniProtKB: Tail tube protein |
-Macromolecule #6: Tape measure protein
| Macromolecule | Name: Tape measure protein / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
| Molecular weight | Theoretical: 92.39343 KDa |
| Sequence | String: MAEPVGDLVV DLSLDAARFD EQMARVRRHF SGTESDAKKT AAVVEQSLSR QALAAQKAGI SVGQYKAAMR MLPAQFTDVA TQLAGGQSP WLILLQQGGQ VKDSFGGMIP MFRGLAGAIT LPMVGATSLA VATGALAYAW YQGNSTLSDF NKTLVLSGNQ A GLTADRML ...String: MAEPVGDLVV DLSLDAARFD EQMARVRRHF SGTESDAKKT AAVVEQSLSR QALAAQKAGI SVGQYKAAMR MLPAQFTDVA TQLAGGQSP WLILLQQGGQ VKDSFGGMIP MFRGLAGAIT LPMVGATSLA VATGALAYAW YQGNSTLSDF NKTLVLSGNQ A GLTADRML VLSRAGQAAG LTFNQTSESL SALVKAGVSG EAQIASISQS VARFSSASGV EVDKVAEAFG KLTTDPTSGL TA MARQFHN VSAEQIAYVA QLQRSGDEAG ALQAANEAAT KGFDDQTRRL KENMGTLETW ADRTARAFKS MWDAVLDIGR PDT AQEMLI KAEAAYKKAD DIWNLRKDDY FVNDEARARY WDDREKARLA LEAARKKAEQ QTQQDKNAQQ QSDTEASRLK YTEE AQKAY ERLQTPLEKY TARQEELNKA LKDGKILQAD YNTLMAAAKK DYEATLKKPK QSSVKVSAGD RQEDSAHAAL LTLQA ELRT LEKHAGANEK ISQQRRDLWK AESQFAVLEE AAQRRQLSAQ EKSLLAHKDE TLEYKRQLAA LGDKVTYQER LNALAQ QAD KFAQQQRAKR AAIDAKSRGL TDRQAEREAT EQRLKEQYGD NPLALNNVMS EQKKTWAAED QLRGNWMAGL KSGWSEW EE SATDSMSQVK SAATQTFDGI AQNMAAMLTG SEQNWRSFTR SVLSMMTEIL LKQAMVGIVG SIGSAIGGAV GGGASASG G TAIQAAAAKF HFATGGFTGT GGKYEPAGIV HRGEFVFTKE ATSRIGVGNL YRLMRGYATG GYVGTPGSMA DSRSQASGT FEQNNHVVIN NDGTNGQIGP AALKAVYDMA RKGARDEIQT QMRDGGLFSG GGR UniProtKB: Tape measure protein |
-Macromolecule #7: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 7 / Number of copies: 3 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)