+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ICP1 Csy-dsDNA complex (form 1) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | IMMUNE SYSTEM-RNA-DNA complex / RNA BINDING PROTEIN/DNA/RNA / RNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Vibrio phage ICP1_2004_A (virus) Vibrio phage ICP1_2004_A (virus) | |||||||||
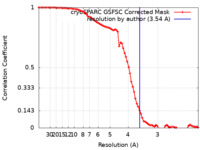
| Method | single particle reconstruction / cryo EM / Resolution: 3.54 Å | |||||||||
 Authors Authors | Zhang LX / Feng Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2024 Journal: Nat Chem Biol / Year: 2024Title: Cas1 mediates the interference stage in a phage-encoded CRISPR-Cas system. Authors: Laixing Zhang / Hao Wang / Jianwei Zeng / Xueli Cao / Zhengyu Gao / Zihe Liu / Feixue Li / Jiawei Wang / Yi Zhang / Maojun Yang / Yue Feng /  Abstract: Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems are prokaryotic adaptive immune systems against invading phages and other mobile genetic elements. Notably, some phages, ...Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems are prokaryotic adaptive immune systems against invading phages and other mobile genetic elements. Notably, some phages, including the Vibrio cholerae-infecting ICP1 (International Center for Diarrheal Disease Research, Bangladesh cholera phage 1), harbor CRISPR-Cas systems to counteract host defenses. Nevertheless, ICP1 Cas8f lacks the helical bundle domain essential for recruitment of helicase-nuclease Cas2/3 during target DNA cleavage and how this system accomplishes the interference stage remains unknown. Here, we found that Cas1, a highly conserved component known to exclusively work in the adaptation stage, also mediates the interference stage through connecting Cas2/3 to the DNA-bound CRISPR-associated complex for antiviral defense (Cascade; CRISPR system yersinia, Csy) of the ICP1 CRISPR-Cas system. A series of structures of Csy, Csy-dsDNA (double-stranded DNA), Cas1-Cas2/3 and Csy-dsDNA-Cas1-Cas2/3 complexes reveal the whole process of Cas1-mediated target DNA cleavage by the ICP1 CRISPR-Cas system. Together, these data support an unprecedented model in which Cas1 mediates the interference stage in a phage-encoded CRISPR-Cas system and the study also sheds light on a unique model of primed adaptation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36831.map.gz emd_36831.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36831-v30.xml emd-36831-v30.xml emd-36831.xml emd-36831.xml | 25.1 KB 25.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36831_fsc.xml emd_36831_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36831.png emd_36831.png | 36.6 KB | ||
| Filedesc metadata |  emd-36831.cif.gz emd-36831.cif.gz | 6.9 KB | ||
| Others |  emd_36831_half_map_1.map.gz emd_36831_half_map_1.map.gz emd_36831_half_map_2.map.gz emd_36831_half_map_2.map.gz | 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36831 http://ftp.pdbj.org/pub/emdb/structures/EMD-36831 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36831 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36831 | HTTPS FTP |
-Related structure data
| Related structure data |  8k28MC  8k0hC  8k0jC  8k0kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36831.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36831.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
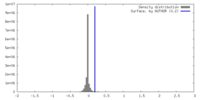
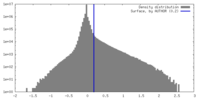
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36831_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
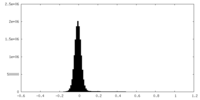
| Density Histograms |
-Half map: #1
| File | emd_36831_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
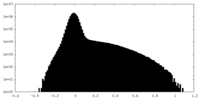
| Density Histograms |
- Sample components
Sample components
-Entire : csy complex
| Entire | Name: csy complex |
|---|---|
| Components |
|
-Supramolecule #1: csy complex
| Supramolecule | Name: csy complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2004_A (virus) Vibrio phage ICP1_2004_A (virus) |
-Macromolecule #1: Csy1
| Macromolecule | Name: Csy1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2004_A (virus) Vibrio phage ICP1_2004_A (virus) |
| Molecular weight | Theoretical: 20.409918 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIKEMIEDFI SKGGLIFTHS GRYTNTNNSC FIFNKNDIGV DTKVDMYTPK SAGIKNEEGE NLWQVLNKAN MFYRIYSGEL GEELQYLLK SCCTAKEDVT TLPQIYFKNG EGYDILVPIG NAHNLISGTE YLWEHKYYNT FTQKLGGSNP QNCTHACNKM R GGFKQFNC TPPQVEDNYN A UniProtKB: Csy1 |
-Macromolecule #2: Csy2
| Macromolecule | Name: Csy2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2004_A (virus) Vibrio phage ICP1_2004_A (virus) |
| Molecular weight | Theoretical: 27.401547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRKFIIVKNV KVDGINAKSS DITVGMPPAT TFCGLGETMS IKTGIVVKAV SYGSVKFEVR GSRFNTSVTK FAWQDRGNGG KANNNSPIQ PKPLADGVFT LCFEVEWEDC AEVLVDKVTN FINTARIAGG TIASFNKPFV KVAKDAEELA SVKNAMMPCY V VVDCGVEV ...String: MRKFIIVKNV KVDGINAKSS DITVGMPPAT TFCGLGETMS IKTGIVVKAV SYGSVKFEVR GSRFNTSVTK FAWQDRGNGG KANNNSPIQ PKPLADGVFT LCFEVEWEDC AEVLVDKVTN FINTARIAGG TIASFNKPFV KVAKDAEELA SVKNAMMPCY V VVDCGVEV NIFEDAVNRK LQPMVNGYKK LEKIVDNKHM RDKFTPAYLA TPTYTMIGYK MVSNVDNFDQ ALWQYGENTK VK TIGGIYN D UniProtKB: Csy2 |
-Macromolecule #3: Csy3
| Macromolecule | Name: Csy3 / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2004_A (virus) Vibrio phage ICP1_2004_A (virus) |
| Molecular weight | Theoretical: 33.399301 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKLKAPAVL AYSRKINPTN ALMFAVNWSD RDNTTAVMVG TKTVAGTQSV RGNPNDADKG NIQTVNFANL PHNKNTLLVK YNVKFVGDV FKAELGGGEY SNTLQTALEN TDFGTLAYRY VYNIAAGRTL WRNRVGAESI ETVITVNDQT FTFSDLLVNE F DEDVDVAE ...String: MTKLKAPAVL AYSRKINPTN ALMFAVNWSD RDNTTAVMVG TKTVAGTQSV RGNPNDADKG NIQTVNFANL PHNKNTLLVK YNVKFVGDV FKAELGGGEY SNTLQTALEN TDFGTLAYRY VYNIAAGRTL WRNRVGAESI ETVITVNDQT FTFSDLLVNE F DEDVDVAE IADMVAGVLS GEGFVTLKVE HYMLLGEGSE VFPSQEFVEN SKLSKQLFDL NGQAAMHDQK IGNAIRTIDT WY EDATTPI AVEPYGSVVR NGVAYRAGNK TDLFTLMDGA VNGKSLTEED QMFVTANLIR GGVFGGGKD UniProtKB: Csy3 |
-Macromolecule #4: Csy4
| Macromolecule | Name: Csy4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2004_A (virus) Vibrio phage ICP1_2004_A (virus) |
| Molecular weight | Theoretical: 18.73668 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYNTISITVV DADDVGVNFV VSKVLSTLHN KGIFNGEVGV TFPRMDKNVG DIITLFSKTG VDRKVLTSTL NTLTDFIHIG KPKEADKVK TYRKVDTKSK GKLIRRCIKR KGVSAETAES LYGNYKGEKC KLPYIVVNSK STGQRFSMFL EECENSEKFN S YGLCIVSC UniProtKB: Csy4 |
-Macromolecule #5: DNA (33-MER)
| Macromolecule | Name: DNA (33-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2004_A (virus) Vibrio phage ICP1_2004_A (virus) |
| Molecular weight | Theoretical: 10.154547 KDa |
| Sequence | String: (DT)(DA)(DA)(DG)(DC)(DA)(DA)(DA)(DG)(DG) (DG)(DT)(DT)(DG)(DA)(DC)(DG)(DA)(DA)(DA) (DG)(DC)(DC)(DC)(DT)(DT)(DT)(DG)(DT) (DC)(DC)(DC)(DT) |
-Macromolecule #6: DNA (27-MER)
| Macromolecule | Name: DNA (27-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2004_A (virus) Vibrio phage ICP1_2004_A (virus) |
| Molecular weight | Theoretical: 8.330393 KDa |
| Sequence | String: (DA)(DG)(DG)(DG)(DA)(DC)(DA)(DA)(DA)(DG) (DG)(DG)(DC)(DT)(DT)(DT)(DC)(DC)(DA)(DT) (DT)(DT)(DA)(DT)(DT)(DT)(DA) |
-Macromolecule #7: RNA (59-MER)
| Macromolecule | Name: RNA (59-MER) / type: rna / ID: 7 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Vibrio phage ICP1_2004_A (virus) Vibrio phage ICP1_2004_A (virus) |
| Molecular weight | Theoretical: 18.701021 KDa |
| Sequence | String: CUUAAAGAGU CAACCCUUUG CUUAUCUUCC CUAUUUAAAU GUUAGCAGCC GCAUAGGCU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)