+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SLC15A4 inhibitor complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | endolysosomal transporter / PROTEIN TRANSPORT-INHIBITOR COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationhistidine transport / mast cell homeostasis / L-histidine transmembrane export from vacuole / peptidoglycan transmembrane transporter activity / L-histidine transmembrane transporter activity / Proton/oligopeptide cotransporters / positive regulation of toll-like receptor 8 signaling pathway / peptidoglycan transport / positive regulation of toll-like receptor 7 signaling pathway / regulation of isotype switching to IgG isotypes ...histidine transport / mast cell homeostasis / L-histidine transmembrane export from vacuole / peptidoglycan transmembrane transporter activity / L-histidine transmembrane transporter activity / Proton/oligopeptide cotransporters / positive regulation of toll-like receptor 8 signaling pathway / peptidoglycan transport / positive regulation of toll-like receptor 7 signaling pathway / regulation of isotype switching to IgG isotypes / SLC15A4:TASL-dependent IRF5 activation / positive regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway / dipeptide import across plasma membrane / positive regulation of toll-like receptor 9 signaling pathway / peptide:proton symporter activity / dipeptide transmembrane transporter activity / regulation of nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway / positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway / endolysosome membrane / positive regulation of innate immune response / specific granule membrane / monoatomic ion transport / protein transport / early endosome membrane / innate immune response / lysosomal membrane / Neutrophil degranulation / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Zhang SS / Chen XD / Xie M | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: A conformation-locking inhibitor of SLC15A4 with TASL proteostatic anti-inflammatory activity. Authors: Andras Boeszoermenyi / Léa Bernaleau / Xudong Chen / Felix Kartnig / Min Xie / Haobo Zhang / Sensen Zhang / Maeva Delacrétaz / Anna Koren / Ann-Katrin Hopp / Vojtech Dvorak / Stefan ...Authors: Andras Boeszoermenyi / Léa Bernaleau / Xudong Chen / Felix Kartnig / Min Xie / Haobo Zhang / Sensen Zhang / Maeva Delacrétaz / Anna Koren / Ann-Katrin Hopp / Vojtech Dvorak / Stefan Kubicek / Daniel Aletaha / Maojun Yang / Manuele Rebsamen / Leonhard X Heinz / Giulio Superti-Furga /    Abstract: Dysregulation of pathogen-recognition pathways of the innate immune system is associated with multiple autoimmune disorders. Due to the intricacies of the molecular network involved, the ...Dysregulation of pathogen-recognition pathways of the innate immune system is associated with multiple autoimmune disorders. Due to the intricacies of the molecular network involved, the identification of pathway- and disease-specific therapeutics has been challenging. Using a phenotypic assay monitoring the degradation of the immune adapter TASL, we identify feeblin, a chemical entity which inhibits the nucleic acid-sensing TLR7/8 pathway activating IRF5 by disrupting the SLC15A4-TASL adapter module. A high-resolution cryo-EM structure of feeblin with SLC15A4 reveals that the inhibitor binds a lysosomal outward-open conformation incompatible with TASL binding on the cytoplasmic side, leading to degradation of TASL. This mechanism of action exploits a conformational switch and converts a target-binding event into proteostatic regulation of the effector protein TASL, interrupting the TLR7/8-IRF5 signaling pathway and preventing downstream proinflammatory responses. Considering that all components involved have been genetically associated with systemic lupus erythematosus and that feeblin blocks responses in disease-relevant human immune cells from patients, the study represents a proof-of-concept for the development of therapeutics against this disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36754.map.gz emd_36754.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36754-v30.xml emd-36754-v30.xml emd-36754.xml emd-36754.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36754.png emd_36754.png | 41.9 KB | ||
| Filedesc metadata |  emd-36754.cif.gz emd-36754.cif.gz | 7.2 KB | ||
| Others |  emd_36754_half_map_1.map.gz emd_36754_half_map_1.map.gz emd_36754_half_map_2.map.gz emd_36754_half_map_2.map.gz | 101.4 MB 101.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36754 http://ftp.pdbj.org/pub/emdb/structures/EMD-36754 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36754 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36754 | HTTPS FTP |
-Related structure data
| Related structure data |  8jzxMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36754.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36754.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8433 Å | ||||||||||||||||||||||||||||||||||||
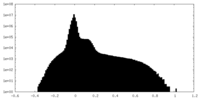
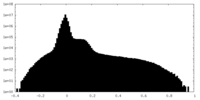
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36754_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
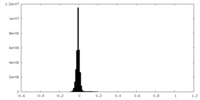
| Density Histograms |
-Half map: #2
| File | emd_36754_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
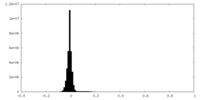
| Density Histograms |
- Sample components
Sample components
-Entire : endolysosomal transporter
| Entire | Name: endolysosomal transporter |
|---|---|
| Components |
|
-Supramolecule #1: endolysosomal transporter
| Supramolecule | Name: endolysosomal transporter / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: SLC15A4-ALFA tag-SLC15A4-twin strep tag fusion protein
| Macromolecule | Name: SLC15A4-ALFA tag-SLC15A4-twin strep tag fusion protein type: protein_or_peptide / ID: 1 Details: SLC15A4 (1-252 residues) + ALFA tag (SRLEEELRRRLTE) + SLC15A4(304-577 resides)+ twin-strep tag sequence Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 62.274246 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEGSGGGAGE RAPLLGARRA AAAAAAAGAF AGRRAACGAV LLTELLERAA FYGITSNLVL FLNGAPFCWE GAQASEALLL FMGLTYLGS PFGGWLADAR LGRARAILLS LALYLLGMLA FPLLAAPATR AALCGSARLL NCTAPGPDAA ARCCSPATFA G LVLVGLGV ...String: MEGSGGGAGE RAPLLGARRA AAAAAAAGAF AGRRAACGAV LLTELLERAA FYGITSNLVL FLNGAPFCWE GAQASEALLL FMGLTYLGS PFGGWLADAR LGRARAILLS LALYLLGMLA FPLLAAPATR AALCGSARLL NCTAPGPDAA ARCCSPATFA G LVLVGLGV ATVKANITPF GADQVKDRGP EATRRFFNWF YWSINLGAIL SLGGIAYIQQ NVSFVTGYAI PTVCVGLAFV VF LCGQSVF ITKPPSRLEE ELRRRLTEGG PFTEEKVEDV KALVKIVPVF LALIPYWTVY FQMQTTYVLQ SLHLRIPEIS NIT TTPHTL PAAWLTMFDA VLILLLIPLK DKLVDPILRR HGLLPSSLKR IAVGMFFVMC SAFAAGILES KRLNLVKEKT INQT IGNVV YHAADLSLWW QVPQYLLIGI SEIFASIAGL EFAYSAAPKS MQSAIMGLFF FFSGVGSFVG SGLLALVSIK AIGWM SSHT DFGNINGCYL NYYFFLLAAI QGATLLLFLI ISVKYDHHRD HQRSRANGVP TSRRAGSLEV LFQGPGGGSG GGSWSH PQF EKGGGSGGGS WSHPQFEK UniProtKB: Solute carrier family 15 member 4, Solute carrier family 15 member 4 |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 4 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #4: 2-(4-ethoxyphenyl)-N-[3-[(2R)-2-methylpiperidin-1-yl]propyl]quino...
| Macromolecule | Name: 2-(4-ethoxyphenyl)-N-[3-[(2R)-2-methylpiperidin-1-yl]propyl]quinoline-4-carboxamide type: ligand / ID: 4 / Number of copies: 2 / Formula: Q09 |
|---|---|
| Molecular weight | Theoretical: 431.57 Da |
| Chemical component information | 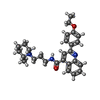 ChemComp-Q09: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)