+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Clobenpropit-bound H4R/Gi complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR-G-protein complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHistamine receptors / histamine receptor activity / neurotransmitter receptor activity / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / regulation of MAPK cascade / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway ...Histamine receptors / histamine receptor activity / neurotransmitter receptor activity / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / regulation of MAPK cascade / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / GDP binding / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of cytosolic calcium ion concentration / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / chemical synaptic transmission / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / inflammatory response / lysosomal membrane / cell division / GTPase activity / synapse / dendrite / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome / nucleoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
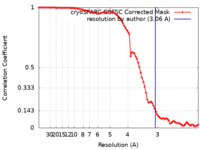
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | He Y / Xia R | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis of ligand recognition and design of antihistamines targeting histamine H receptor. Authors: Ruixue Xia / Shuang Shi / Zhenmei Xu / Henry F Vischer / Albert D Windhorst / Yu Qian / Yaning Duan / Jiale Liang / Kai Chen / Anqi Zhang / Changyou Guo / Rob Leurs / Yuanzheng He /   Abstract: The histamine H receptor (HR) plays key role in immune cell function and is a highly valued target for treating allergic and inflammatory diseases. However, structural information of HR remains ...The histamine H receptor (HR) plays key role in immune cell function and is a highly valued target for treating allergic and inflammatory diseases. However, structural information of HR remains elusive. Here, we report four cryo-EM structures of HR/G complexes, with either histamine or synthetic agonists clobenpropit, VUF6884 and clozapine bound. Combined with mutagenesis, ligand binding and functional assays, the structural data reveal a distinct ligand binding mode where D94 and a π-π network determine the orientation of the positively charged group of ligands, while E182, located at the opposite end of the ligand binding pocket, plays a key role in regulating receptor activity. The structural insight into HR ligand binding allows us to identify mutants at E182 for which the agonist clobenpropit acts as an inverse agonist and to correctly predict inverse agonism of a closely related analog with nanomolar potency. Together with the findings regarding receptor activation and G engagement, we establish a framework for understanding HR signaling and provide a rational basis for designing novel antihistamines targeting HR. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36716.map.gz emd_36716.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36716-v30.xml emd-36716-v30.xml emd-36716.xml emd-36716.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36716_fsc.xml emd_36716_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_36716.png emd_36716.png | 81 KB | ||
| Filedesc metadata |  emd-36716.cif.gz emd-36716.cif.gz | 6.6 KB | ||
| Others |  emd_36716_half_map_1.map.gz emd_36716_half_map_1.map.gz emd_36716_half_map_2.map.gz emd_36716_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36716 http://ftp.pdbj.org/pub/emdb/structures/EMD-36716 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36716 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36716 | HTTPS FTP |
-Validation report
| Summary document |  emd_36716_validation.pdf.gz emd_36716_validation.pdf.gz | 927 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36716_full_validation.pdf.gz emd_36716_full_validation.pdf.gz | 926.5 KB | Display | |
| Data in XML |  emd_36716_validation.xml.gz emd_36716_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_36716_validation.cif.gz emd_36716_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36716 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36716 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36716 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36716 | HTTPS FTP |
-Related structure data
| Related structure data |  8jxxMC  8jxtC  8jxvC  8jxwC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36716.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36716.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36716_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36716_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GPCR/G-protein complex
| Entire | Name: GPCR/G-protein complex |
|---|---|
| Components |
|
-Supramolecule #1: GPCR/G-protein complex
| Supramolecule | Name: GPCR/G-protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 130 MDa |
-Macromolecule #1: Histamine H4 receptor
| Macromolecule | Name: Histamine H4 receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.543766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPDTNSTINL SLSTRVTLAF FMSLVAFAIM LGNALVILAF VVDKNLRHRS SYFFLNLAIS DFFVGVISIP LYIPHTLFEW DFGKEICVF WLTTDYLLCT ASVYNIVLIS YDRYLSVSNA VSYRTQHTGV LKIVTLMVAV WVLAFLVNGP MILVSESWKD E GSECEPGF ...String: MPDTNSTINL SLSTRVTLAF FMSLVAFAIM LGNALVILAF VVDKNLRHRS SYFFLNLAIS DFFVGVISIP LYIPHTLFEW DFGKEICVF WLTTDYLLCT ASVYNIVLIS YDRYLSVSNA VSYRTQHTGV LKIVTLMVAV WVLAFLVNGP MILVSESWKD E GSECEPGF FSEWYILAIT SFLEFVIPVI LVAYFNMNIY WSLWKRDHLS RCQSHPGLTA VSSNICGHSF RGRLSSRRSL SA STEVPAS FHSERQRRKS SLMFSSRTKM NSNTIASKMG SFSQSDSVAL HQREHVELLR ARRLAKSLAI LLGVFAVCWA PYS LFTIVL SFYSSATGPK SVWYRIAFWL QWFNSFVNPL LYPLCHKRFQ KAFLKIFCIK KQPLPSQHSR SVSS UniProtKB: Histamine H4 receptor |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.445059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.277299 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL |
-Macromolecule #6: 3-(1~{H}-imidazol-4-yl)propyl ~{N}'-[(4-chlorophenyl)methyl]carba...
| Macromolecule | Name: 3-(1~{H}-imidazol-4-yl)propyl ~{N}'-[(4-chlorophenyl)methyl]carbamimidothioate type: ligand / ID: 6 / Number of copies: 1 / Formula: VCL |
|---|---|
| Molecular weight | Theoretical: 308.83 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)