+検索条件
-Structure paper
| タイトル | Structural basis of ligand recognition and design of antihistamines targeting histamine H receptor. |
|---|---|
| ジャーナル・号・ページ | Nat Commun, Vol. 15, Issue 1, Page 2493, Year 2024 |
| 掲載日 | 2024年3月20日 |
 著者 著者 | Ruixue Xia / Shuang Shi / Zhenmei Xu / Henry F Vischer / Albert D Windhorst / Yu Qian / Yaning Duan / Jiale Liang / Kai Chen / Anqi Zhang / Changyou Guo / Rob Leurs / Yuanzheng He /   |
| PubMed 要旨 | The histamine H receptor (HR) plays key role in immune cell function and is a highly valued target for treating allergic and inflammatory diseases. However, structural information of HR remains ...The histamine H receptor (HR) plays key role in immune cell function and is a highly valued target for treating allergic and inflammatory diseases. However, structural information of HR remains elusive. Here, we report four cryo-EM structures of HR/G complexes, with either histamine or synthetic agonists clobenpropit, VUF6884 and clozapine bound. Combined with mutagenesis, ligand binding and functional assays, the structural data reveal a distinct ligand binding mode where D94 and a π-π network determine the orientation of the positively charged group of ligands, while E182, located at the opposite end of the ligand binding pocket, plays a key role in regulating receptor activity. The structural insight into HR ligand binding allows us to identify mutants at E182 for which the agonist clobenpropit acts as an inverse agonist and to correctly predict inverse agonism of a closely related analog with nanomolar potency. Together with the findings regarding receptor activation and G engagement, we establish a framework for understanding HR signaling and provide a rational basis for designing novel antihistamines targeting HR. |
 リンク リンク |  Nat Commun / Nat Commun /  PubMed:38509098 / PubMed:38509098 /  PubMed Central PubMed Central |
| 手法 | EM (単粒子) |
| 解像度 | 3.01 - 3.21 Å |
| 構造データ | EMDB-36712, PDB-8jxt: EMDB-36714, PDB-8jxv: EMDB-36715, PDB-8jxw: EMDB-36716, PDB-8jxx: |
| 化合物 | 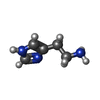 ChemComp-HSM:  ChemComp-PO4: 
ChemComp-VBU: 
ChemComp-VCF: 
ChemComp-VCL: |
| 由来 |
|
 キーワード キーワード | MEMBRANE PROTEIN / GPCR-G-protein complex |
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア 万見文献について
万見文献について











 homo sapiens (ヒト)
homo sapiens (ヒト)