+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3671 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
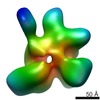
| Title | Open dimer of human ATM (Ataxia telangiectasia mutated) | |||||||||
 Map data Map data | Open dimer of human ATM (Ataxia telangiectasia mutated) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.5 Å | |||||||||
 Authors Authors | Baretic D / Pollard HK / Fisher DI / Johnson CM / Santhanam B / Truman CM / Kouba T / Fersht AR / Phillips C | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2017 Journal: Sci Adv / Year: 2017Title: Structures of closed and open conformations of dimeric human ATM. Authors: Domagoj Baretić / Hannah K Pollard / David I Fisher / Christopher M Johnson / Balaji Santhanam / Caroline M Truman / Tomas Kouba / Alan R Fersht / Christopher Phillips / Roger L Williams /  Abstract: ATM (ataxia-telangiectasia mutated) is a phosphatidylinositol 3-kinase-related protein kinase (PIKK) best known for its role in DNA damage response. ATM also functions in oxidative stress response, ...ATM (ataxia-telangiectasia mutated) is a phosphatidylinositol 3-kinase-related protein kinase (PIKK) best known for its role in DNA damage response. ATM also functions in oxidative stress response, insulin signaling, and neurogenesis. Our electron cryomicroscopy (cryo-EM) suggests that human ATM is in a dynamic equilibrium between closed and open dimers. In the closed state, the PIKK regulatory domain blocks the peptide substrate-binding site, suggesting that this conformation may represent an inactive or basally active enzyme. The active site is held in this closed conformation by interaction with a long helical hairpin in the TRD3 (tetratricopeptide repeats domain 3) domain of the symmetry-related molecule. The open dimer has two protomers with only a limited contact interface, and it lacks the intermolecular interactions that block the peptide-binding site in the closed dimer. This suggests that the open conformation may be more active. The ATM structure shows the detailed topology of the regulator-interacting N-terminal helical solenoid. The ATM conformational dynamics shown by the structures represent an important step in understanding the enzyme regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3671.map.gz emd_3671.map.gz | 83.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3671-v30.xml emd-3671-v30.xml emd-3671.xml emd-3671.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3671_fsc.xml emd_3671_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3671.png emd_3671.png | 66.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3671 http://ftp.pdbj.org/pub/emdb/structures/EMD-3671 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3671 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3671 | HTTPS FTP |
-Related structure data
| Related structure data |  3668C  3669C  3670C  3672C  3673C  5np0C  5np1C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3671.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3671.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Open dimer of human ATM (Ataxia telangiectasia mutated) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.43 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dimeric human ATM (Ataxia telangiectasia mutated) kinase
| Entire | Name: Dimeric human ATM (Ataxia telangiectasia mutated) kinase |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric human ATM (Ataxia telangiectasia mutated) kinase
| Supramolecule | Name: Dimeric human ATM (Ataxia telangiectasia mutated) kinase type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: ATM was produced and imaged with the FLAG tag at the N-terminus |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Experimental: 705 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: Expi293F Homo sapiens (human) / Recombinant cell: Expi293F |
-Macromolecule #1: human ATM (Ataxia Telangiectasia mutated)
| Macromolecule | Name: human ATM (Ataxia Telangiectasia mutated) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKH MS LVLNDLL ICCRQLEHDR ATE RKKEVE KFKRLIRDPE TIKH LDRHS DSKQGKYLNW DAVFR FLQK YIQKETECLR IAKPNV SAS TQASRQKKMQ EISSLVK YF IKCANRRAPR LKCQELLN Y IMDTVKDSSN GAIYGADCS NILLKDILSV ...String: MDYKDDDDKH MS LVLNDLL ICCRQLEHDR ATE RKKEVE KFKRLIRDPE TIKH LDRHS DSKQGKYLNW DAVFR FLQK YIQKETECLR IAKPNV SAS TQASRQKKMQ EISSLVK YF IKCANRRAPR LKCQELLN Y IMDTVKDSSN GAIYGADCS NILLKDILSV RKYWCEISQQ QWLELFSVY FRLYLKPSQD V HRVLVARI IHAVTKGCCS QT DGLNSKF LDFFSKAIQC ARQ EKSSSG LNHILAALTI FLKT LAVNF RIRVCELGDE ILPTL LYIW TQHRLNDSLK EVIIEL FQL QIYIHHPKGA KTQEKGA YE STKWRSILYN LYDLLVNE I SHIGSRGKYS SGFRNIAVK ENLIELMADI CHQVFNEDTR SLEISQSYT TTQRESSDYS V PCKRKKIE LGWEVIKDHL QK SQNDFDL VPWLQIATQL ISK YPASLP NCELSPLLMI LSQL LPQQR HGERTPYVLR CLTEV ALCQ DKRSNLESSQ KSDLLK LWN KIWCITFRGI SSEQIQA EN FGLLGAIIQG SLVEVDRE F WKLFTGSACR PSCPAVCCL TLALTTSIVP GTVKMGIEQN MCEVNRSFS LKESIMKWLL F YQLEGDLE NSTEVPPILH SN FPHLVLE KILVSLTMKN CKA AMNFFQ SVPECEHHQK DKEE LSFSE VEELFLQTTF DKMDF LTIV RECGIEKHQS SIGFSV HQN LKESLDRCLL GLSEQLL NN YSSEITNSET LVRCSRLL V GVLGCYCYMG VIAEEEAYK SELFQKAKSL MQCAGESITL FKNKTNEEF RIGSLRNMMQ L CTRCLSNC TKKSPNKIAS GF FLRLLTS KLMNDIADIC KSL ASFIKK PFDRGEVESM EDDT NGNLM EVEDQSSMNL FNDYP DSSV SDANEPGESQ STIGAI NPL AEEYLSKQDL LFLDMLK FL CLCVTTAQTN TVSFRAAD I RRKLLMLIDS STLEPTKSL HLHMYLMLLK ELPGEEYPLP MEDVLELLK PLSNVCSLYR R DQDVCKTI LNHVLHVVKN LG QSNMDSE NTRDAQGQFL TVI GAFWHL TKERKYIFSV RMAL VNCLK TLLEADPYSK WAILN VMGK DFPVNEVFTQ FLADNH HQV RMLAAESINR LFQDTKG DS SRLLKALPLK LQQTAFEN A YLKAQEGMRE MSHSAENPE TLDEIYNRKS VLLTLIAVVL SCSPICEKQ ALFALCKSVK E NGLEPHLV KKVLEKVSET FG YRRLEDF MASHLDYLVL EWL NLQDTE YNLSSFPFIL LNYT NIEDF YRSCYKVLIP HLVIR SHFD EVKSIANQIQ EDWKSL LTD CFPKILVNIL PYFAYEG TR DSGMAQQRET ATKVYDML K SENLLGKQID HLFISNLPE IVVELLMTLH EPANSSASQS TDLCDFSGD LDPAPNPPHF P SHVIKATF AYISNCHKTK LK SILEILS KSPDSYQKIL LAI CEQAAE TNNVYKKHRI LKIY HLFVS LLLKDIKSGL GGAWA FVLR DVIYTLIHYI NQRPSC IMD VSLRSFSLCC DLLSQVC QT AVTYCKDALE NHLHVIVG T LIPLVYEQVE VQKQVLDLL KYLVIDNKDN ENLYITIKLL DPFPDHVVF KDLRITQQKI K YSRGPFSL LEEINHFLSV SV YDALPLT RLEGLKDLRR QLE LHKDQM VDIMRASQDN PQDG IMVKL VVNLLQLSKM AINHT GEKE VLEAVGSCLG EVGPID FST IAIQHSKDAS YTKALKL FE DKELQWTFIM LTYLNNTL V EDCVKVRSAA VTCLKNILA TKTGHSFWEI YKMTTDPMLA YLQPFRTSR KKFLEVPRFD K ENPFEGLD DINLWIPLSE NH DIWIKTL TCAFLDSGGT KCE ILQLLK PMCEVKTDFC QTVL PYLIH DILLQDTNES WRNLL STHV QGFFTSCLRH FSQTSR STT PANLDSESEH FFRCCLD KK SQRTMLAVVD YMRRQKRP S SGTIFNDAFW LDLNYLEVA KVAQSCAAHF TALLYAEIYA DKKSMDDQE KRSLAFEEGS Q STTISSLS EKSKEETGIS LQ DLLLEIY RSIGEPDSLY GCG GGKMLQ PITRLRTYEH EAMW GKALV TYDLETAIPS STRQA GIIQ ALQNLGLCHI LSVYLK GLD YENKDWCPEL EELHYQA AW RNMQWDHCTS VSKEVEGT S YHESLYNALQ SLRDREFST FYESLKYARV KEVEEMCKRS LESVYSLYP TLSRLQAIGE L ESIGELFS RSVTHRQLSE VY IKWQKHS QLLKDSDFSF QEP IMALRT VILEILMEKE MDNS QRECI KDILTKHLVE LSILA RTFK NTQLPERAIF QIKQYN SVS CGVSEWQLEE AQVFWAK KE QSLALSILKQ MIKKLDAS C AANNPSLKLT YTECLRVCG NWLAETCLEN PAVIMQTYLE KAVEVAGNY DGESSDELRN G KMKAFLSL ARFSDTQYQR IE NYMKSSE FENKQALLKR AKE EVGLLR EHKIQTNRYT VKVQ RELEL DELALRALKE DRKRF LCKA VENYINCLLS GEEHDM WVF RLCSLWLENS GVSEVNG MM KRDGMKIPTY KFLPLMYQ L AARMGTKMMG GLGFHEVLN NLISRISMDH PHHTLFIILA LANANRDEF LTKPEVARRS R ITKNVPKQ SSQLDEDRTE AA NRIICTI RSRRPQMVRS VEA LCDAYI ILANLDATQW KTQR KGINI PADQPITKLK NLEDV VVPT MEIKVDHTGE YGNLVT IQS FKAEFRLAGG VNLPKII DC VGSDGKERRQ LVKGRDDL R QDAVMQQVFQ MCNTLLQRN TETRKRKLTI CTYKVVPLSQ RSGVLEWCT GTVPIGEFLV N NEDGAHKR YRPNDFSAFQ CQ KKMMEVQ KKSFEEKYEV FMD VCQNFQ PVFRYFCMEK FLDP AIWFE KRLAYTRSVA TSSIV GYIL GLGDRHVQNI LINEQS AEL VHIDLGVAFE QGKILPT PE TVPFRLTRDI VDGMGITG V EGVFRRCCEK TMEVMRNSQ ETLLTIVEVL LYDPLFDWTM NPLKALYLQ QRPEDETELH P TLNADDQE CKRNLSDIDQ SF NKVAERV LMRLQEKLKG VEE GTVLSV GGQVNLLIQQ AIDP KNLSR LFPGWKAWV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation #1
Sample preparation #1
| Preparation ID | 1 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Concentration | 0.6 mg/mL | ||||||||||||||||||
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Film type ID: 1 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER Details: 3 uL of sample/grid blotted for 12 s before plunge-freezing. | ||||||||||||||||||
| Details | The sample was purified by anti-FLAG affinity chromatography followed by overnight dialysis and a final size-exclusion chromatography. |
- Sample preparation #2
Sample preparation #2
| Preparation ID | 2 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Film type ID: 1 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER Details: 3 uL of sample/grid blotted for 12 s before plunge-freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.15 K |
| Specialist optics | Energy filter - Name: GIF / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-20 / Number grids imaged: 4 / Number real images: 2720 / Average exposure time: 0.8 sec. / Average electron dose: 2.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 35714 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 97902 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















