[English] 日本語
 Yorodumi
Yorodumi- EMDB-3651: Cryo-EM asymmetric reconstruction of haemoglobin at 3.6 A determi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3651 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM asymmetric reconstruction of haemoglobin at 3.6 A determined with the Volta phase plate (subset of particles) | |||||||||
 Map data Map data | Postprocessed asymmetric masked map of haemoglobin dataset with 76150 particles | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Khoshouei M / Radjainia M / Baumeister W / Danev R | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Cryo-EM structure of haemoglobin at 3.2 Å determined with the Volta phase plate. Authors: Maryam Khoshouei / Mazdak Radjainia / Wolfgang Baumeister / Radostin Danev /   Abstract: With the advent of direct electron detectors, the perspectives of cryo-electron microscopy (cryo-EM) have changed in a profound way. These cameras are superior to previous detectors in coping with ...With the advent of direct electron detectors, the perspectives of cryo-electron microscopy (cryo-EM) have changed in a profound way. These cameras are superior to previous detectors in coping with the intrinsically low contrast and beam-induced motion of radiation-sensitive organic materials embedded in amorphous ice, and hence they have enabled the structure determination of many macromolecular assemblies to atomic or near-atomic resolution. Nevertheless, there are still limitations and one of them is the size of the target structure. Here, we report the use of a Volta phase plate in determining the structure of human haemoglobin (64 kDa) at 3.2 Å. Our results demonstrate that this method can be applied to complexes that are significantly smaller than those previously studied by conventional defocus-based approaches. Cryo-EM is now close to becoming a fast and cost-effective alternative to crystallography for high-resolution protein structure determination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3651.map.gz emd_3651.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3651-v30.xml emd-3651-v30.xml emd-3651.xml emd-3651.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3651.png emd_3651.png | 76.2 KB | ||
| Others |  emd_3651_half_map_1.map.gz emd_3651_half_map_1.map.gz emd_3651_half_map_2.map.gz emd_3651_half_map_2.map.gz | 2.8 MB 2.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3651 http://ftp.pdbj.org/pub/emdb/structures/EMD-3651 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3651 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3651 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3651.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3651.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed asymmetric masked map of haemoglobin dataset with 76150 particles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
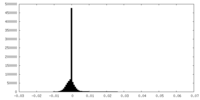
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half1 map from haemoglobin dataset with 76150 particles
| File | emd_3651_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half1 map from haemoglobin dataset with 76150 particles | ||||||||||||
| Projections & Slices |
| ||||||||||||
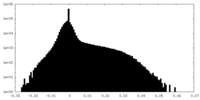
| Density Histograms |
-Half map: Half2 map from haemoglobin dataset with 76150 particles
| File | emd_3651_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half2 map from haemoglobin dataset with 76150 particles | ||||||||||||
| Projections & Slices |
| ||||||||||||
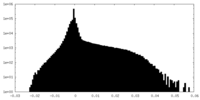
| Density Histograms |
- Sample components
Sample components
-Entire : Hemoglobin
| Entire | Name: Hemoglobin |
|---|---|
| Components |
|
-Supramolecule #1: Hemoglobin
| Supramolecule | Name: Hemoglobin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 76150 |
|---|---|
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)