+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3628 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA polymerase II-Paf1C-TFIIS-B | |||||||||||||||
 Map data Map data | Non-B factor sharpened, locally filtered map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |   | |||||||||||||||
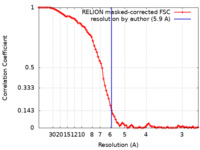
| Method | single particle reconstruction / cryo EM / Resolution: 5.9 Å | |||||||||||||||
 Authors Authors | Xu Y / Cramer P | |||||||||||||||
| Funding support |  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Authors: Youwei Xu / Carrie Bernecky / Chung-Tien Lee / Kerstin C Maier / Björn Schwalb / Dimitry Tegunov / Jürgen M Plitzko / Henning Urlaub / Patrick Cramer /  Abstract: The conserved polymerase-associated factor 1 complex (Paf1C) plays multiple roles in chromatin transcription and genomic regulation. Paf1C comprises the five subunits Paf1, Leo1, Ctr9, Cdc73 and ...The conserved polymerase-associated factor 1 complex (Paf1C) plays multiple roles in chromatin transcription and genomic regulation. Paf1C comprises the five subunits Paf1, Leo1, Ctr9, Cdc73 and Rtf1, and binds to the RNA polymerase II (Pol II) transcription elongation complex (EC). Here we report the reconstitution of Paf1C from Saccharomyces cerevisiae, and a structural analysis of Paf1C bound to a Pol II EC containing the elongation factor TFIIS. Cryo-electron microscopy and crosslinking data reveal that Paf1C is highly mobile and extends over the outer Pol II surface from the Rpb2 to the Rpb3 subunit. The Paf1-Leo1 heterodimer and Cdc73 form opposite ends of Paf1C, whereas Ctr9 bridges between them. Consistent with the structural observations, the initiation factor TFIIF impairs Paf1C binding to Pol II, whereas the elongation factor TFIIS enhances it. We further show that Paf1C is globally required for normal mRNA transcription in yeast. These results provide a three-dimensional framework for further analysis of Paf1C function in transcription through chromatin. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3628.map.gz emd_3628.map.gz | 48.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3628-v30.xml emd-3628-v30.xml emd-3628.xml emd-3628.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3628_fsc.xml emd_3628_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_3628.png emd_3628.png | 58 KB | ||
| Masks |  emd_3628_msk_1.map emd_3628_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Others |  emd_3628_half_map_1.map.gz emd_3628_half_map_1.map.gz emd_3628_half_map_2.map.gz emd_3628_half_map_2.map.gz | 40.8 MB 40.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3628 http://ftp.pdbj.org/pub/emdb/structures/EMD-3628 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3628 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3628 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3628.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3628.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Non-B factor sharpened, locally filtered map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_3628_msk_1.map emd_3628_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_3628_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_3628_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RNA polymerase II-Paf1C-TFIIS-B
| Entire | Name: RNA polymerase II-Paf1C-TFIIS-B |
|---|---|
| Components |
|
-Supramolecule #1: RNA polymerase II-Paf1C-TFIIS-B
| Supramolecule | Name: RNA polymerase II-Paf1C-TFIIS-B / type: complex / ID: 1 / Parent: 0 / Details: Elongation complex |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 900 KDa |
-Supramolecule #2: Paf1C
| Supramolecule | Name: Paf1C / type: complex / ID: 2 / Parent: 1 / Details: Ctr9 C-terminal truncation |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 340 KDa |
-Supramolecule #3: RNA polymerase II
| Supramolecule | Name: RNA polymerase II / type: complex / ID: 3 / Parent: 1 / Details: 12 subunits |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 500 KDa |
-Supramolecule #4: TFIIS
| Supramolecule | Name: TFIIS / type: complex / ID: 4 / Parent: 1 / Details: TFIIS (D290A, E291A) inactive mutant |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 35 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil Cu R3.5/1 and Cu R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 4 micro-L of sample was placed onto Quantifoil Cu R3.5/1 and Cu R2/1 glow-discharged 200 mesh holey carbon grids, which were then blotted for 8.5 s with blot force 13 to remove the excess ...Details: 4 micro-L of sample was placed onto Quantifoil Cu R3.5/1 and Cu R2/1 glow-discharged 200 mesh holey carbon grids, which were then blotted for 8.5 s with blot force 13 to remove the excess solution before they were flash-frozen in liquid ethane.. | ||||||||||||||||||
| Details | Gradient fixation |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Detector mode: SUPER-RESOLUTION / #0 - Number grids imaged: 1 / #0 - Number real images: 595 / #0 - Average exposure time: 0.4 sec. / #0 - Average electron dose: 1.22 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #1 - Detector mode: SUPER-RESOLUTION / #1 - Number grids imaged: 1 / #1 - Number real images: 2146 / #1 - Average exposure time: 0.4 sec. / #1 - Average electron dose: 0.93 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 37000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)